Probe CUST_46284_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46284_PI426222305 | JHI_St_60k_v1 | DMT400048796 | GCTACCGTTCTTGCCCAGACAAGGTATATAGGGAACTCGAAACAAATGACTCAAATCTTG |
All Microarray Probes Designed to Gene DMG402018962
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46284_PI426222305 | JHI_St_60k_v1 | DMT400048796 | GCTACCGTTCTTGCCCAGACAAGGTATATAGGGAACTCGAAACAAATGACTCAAATCTTG |
Microarray Signals from CUST_46284_PI426222305
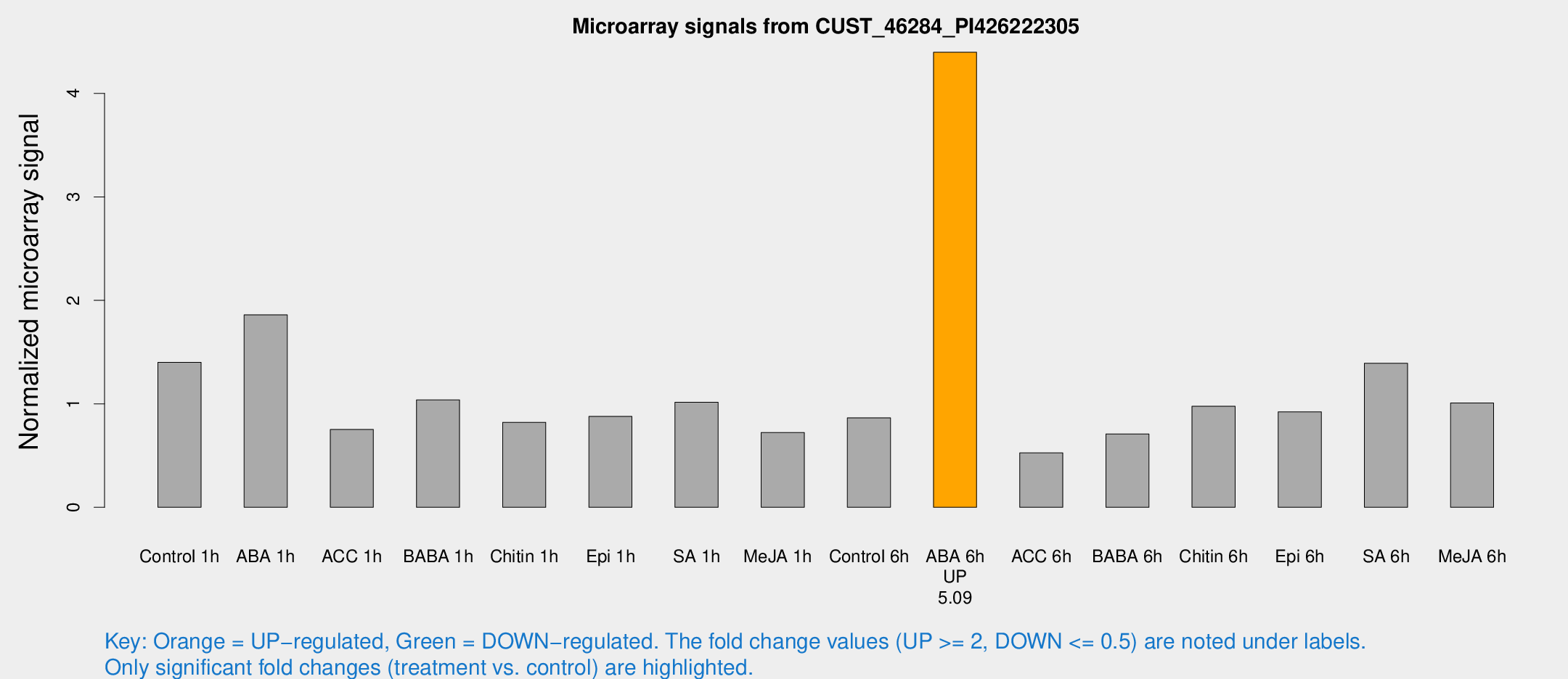
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 759.185 | 85.0096 | 1.40138 | 0.0811486 |
| ABA 1h | 927.616 | 193.203 | 1.86003 | 0.364024 |
| ACC 1h | 518.182 | 188.623 | 0.752363 | 0.405975 |
| BABA 1h | 565.021 | 123.168 | 1.03813 | 0.149418 |
| Chitin 1h | 415.077 | 95.3811 | 0.821686 | 0.145147 |
| Epi 1h | 416.592 | 63.4581 | 0.877921 | 0.137609 |
| SA 1h | 568.882 | 75.3806 | 1.01577 | 0.0967516 |
| Me-JA 1h | 320.626 | 42.2847 | 0.722596 | 0.0424612 |
| Control 6h | 488.541 | 113.409 | 0.864257 | 0.153214 |
| ABA 6h | 2589.62 | 449.678 | 4.39804 | 0.672017 |
| ACC 6h | 323.576 | 19.2185 | 0.525874 | 0.0879349 |
| BABA 6h | 433.679 | 63.861 | 0.708816 | 0.12214 |
| Chitin 6h | 560.909 | 56.6462 | 0.976242 | 0.101803 |
| Epi 6h | 579.218 | 113.413 | 0.922162 | 0.121208 |
| SA 6h | 787.953 | 215.024 | 1.39168 | 0.280916 |
| Me-JA 6h | 570.43 | 142.067 | 1.00755 | 0.169226 |
Source Transcript PGSC0003DMT400048796 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G78950.1 | +1 | 0.0 | 1231 | 572/759 (75%) | Terpenoid cyclases family protein | chr1:29684558-29688673 REVERSE LENGTH=759 |
