Probe CUST_46166_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46166_PI426222305 | JHI_St_60k_v1 | DMT400095756 | ATTGGTTGTTTTGTGACTCACTGTGGATGGAATTCAACTTTGGAGAGCATAGCATCTGAG |
All Microarray Probes Designed to Gene DMG400045327
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46166_PI426222305 | JHI_St_60k_v1 | DMT400095756 | ATTGGTTGTTTTGTGACTCACTGTGGATGGAATTCAACTTTGGAGAGCATAGCATCTGAG |
Microarray Signals from CUST_46166_PI426222305
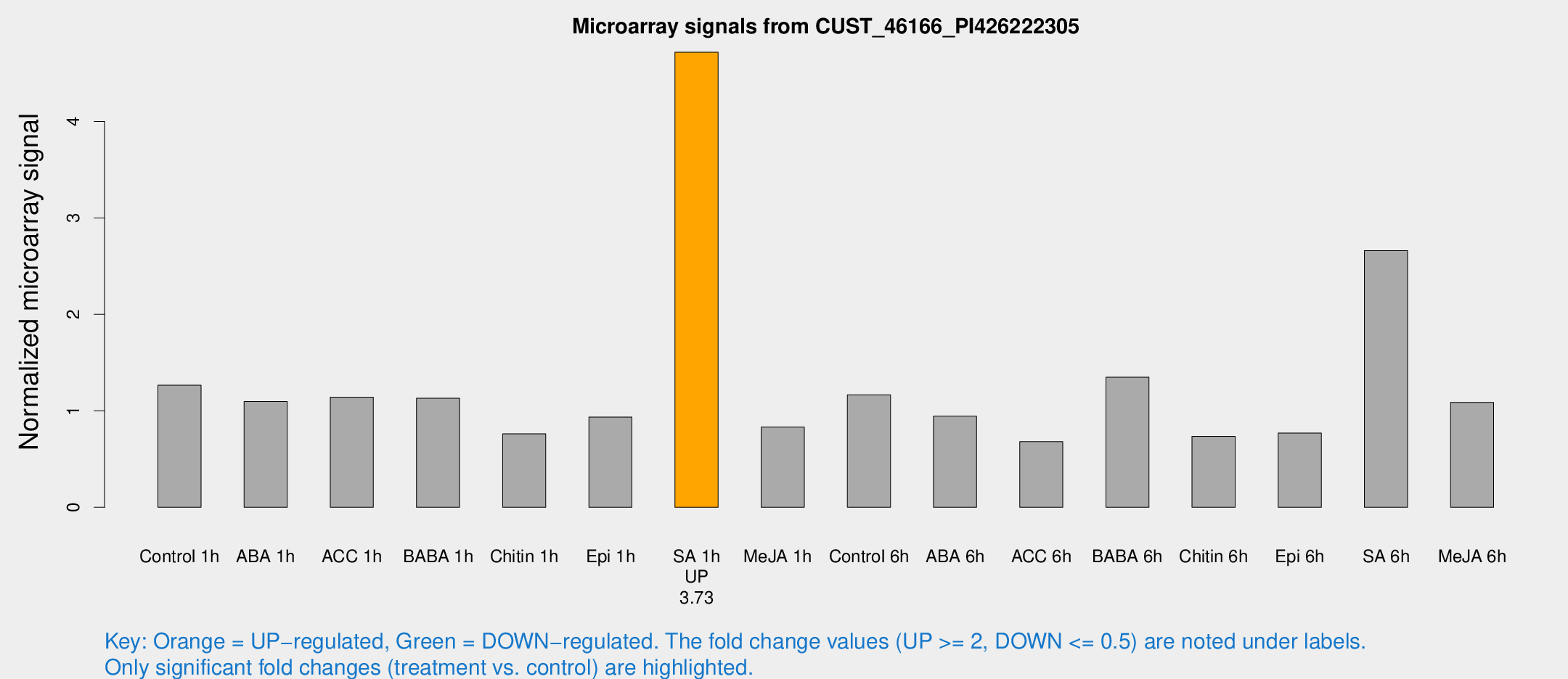
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1983.36 | 423.74 | 1.26551 | 0.190997 |
| ABA 1h | 1504.54 | 281.832 | 1.09584 | 0.189361 |
| ACC 1h | 1857.61 | 432.527 | 1.14186 | 0.211771 |
| BABA 1h | 1690.03 | 296.693 | 1.13094 | 0.104848 |
| Chitin 1h | 1083.76 | 256.979 | 0.761472 | 0.121649 |
| Epi 1h | 1219.67 | 103.606 | 0.935067 | 0.0540565 |
| SA 1h | 7366.97 | 932.097 | 4.71738 | 0.300544 |
| Me-JA 1h | 1051.1 | 203.653 | 0.831073 | 0.091362 |
| Control 6h | 1855.05 | 440.138 | 1.16573 | 0.187438 |
| ABA 6h | 1574.65 | 337.284 | 0.945777 | 0.150624 |
| ACC 6h | 1182.95 | 127.769 | 0.681173 | 0.12075 |
| BABA 6h | 2289.88 | 268.505 | 1.34905 | 0.127764 |
| Chitin 6h | 1184.15 | 115.366 | 0.735902 | 0.0622399 |
| Epi 6h | 1305.62 | 86.9777 | 0.770279 | 0.0546864 |
| SA 6h | 4346.95 | 1232.99 | 2.6614 | 0.600301 |
| Me-JA 6h | 1706.17 | 386.602 | 1.08804 | 0.2231 |
Source Transcript PGSC0003DMT400095756 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G05560.1 | +1 | 4e-25 | 100 | 54/135 (40%) | UDP-glucosyltransferase 75B1 | chr1:1645674-1647083 REVERSE LENGTH=469 |
