Probe CUST_46087_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46087_PI426222305 | JHI_St_60k_v1 | DMT400018519 | TCTTGGAGAGGTCTTGCACTGCTGAGTTCTCTGGTTTTCTTCTCTACAAGGAACTTGGAA |
All Microarray Probes Designed to Gene DMG400007188
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46087_PI426222305 | JHI_St_60k_v1 | DMT400018519 | TCTTGGAGAGGTCTTGCACTGCTGAGTTCTCTGGTTTTCTTCTCTACAAGGAACTTGGAA |
Microarray Signals from CUST_46087_PI426222305
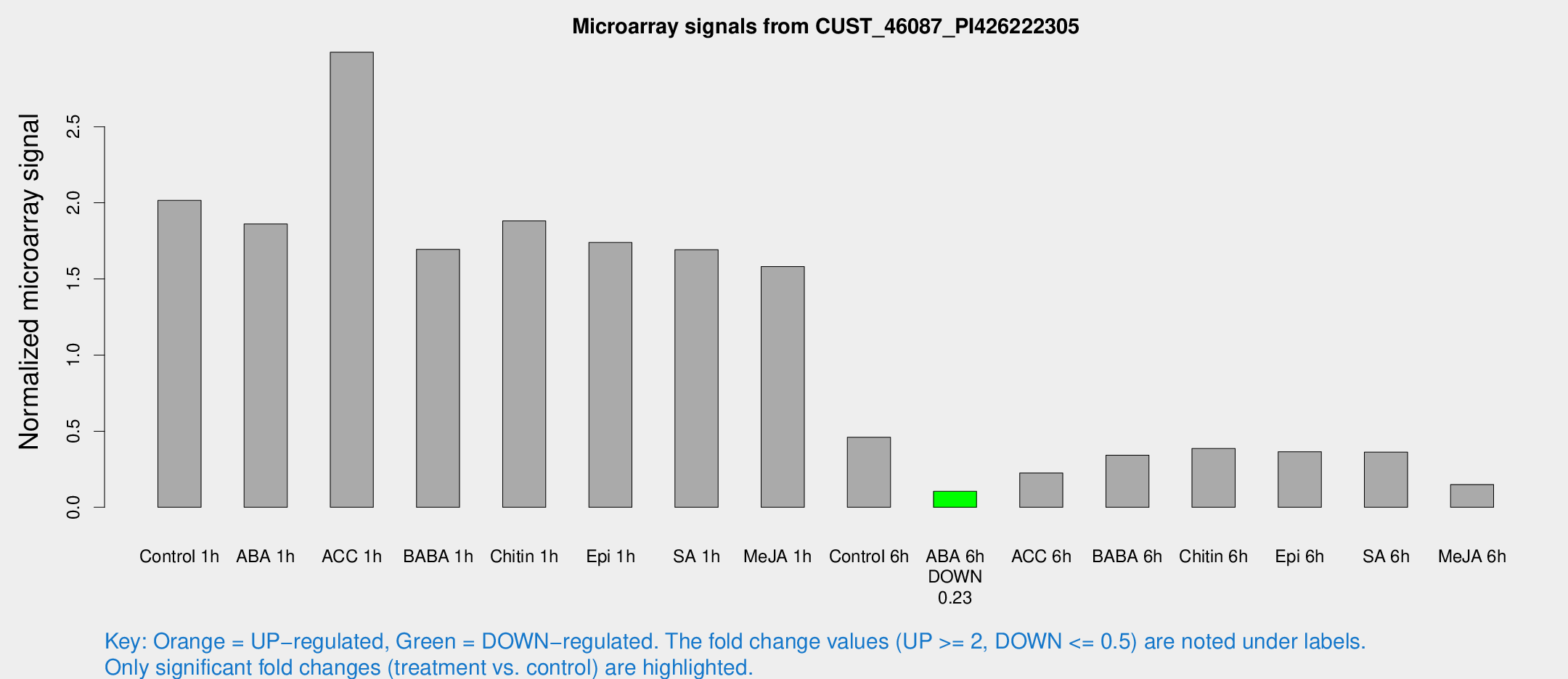
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 13962.3 | 3015.47 | 2.01545 | 0.301096 |
| ABA 1h | 10977.9 | 925.469 | 1.86156 | 0.18542 |
| ACC 1h | 20871.7 | 3239.98 | 2.98916 | 0.630687 |
| BABA 1h | 11006.6 | 1422.01 | 1.69404 | 0.0978072 |
| Chitin 1h | 11580.6 | 2218.71 | 1.881 | 0.353608 |
| Epi 1h | 10022.1 | 791.801 | 1.7394 | 0.100426 |
| SA 1h | 11622.2 | 1220.18 | 1.69196 | 0.122057 |
| Me-JA 1h | 8648.87 | 962.561 | 1.58138 | 0.165539 |
| Control 6h | 3527.7 | 1108.72 | 0.459641 | 0.159917 |
| ABA 6h | 741.009 | 43.0217 | 0.105171 | 0.00927654 |
| ACC 6h | 1946.43 | 670.612 | 0.225295 | 0.058494 |
| BABA 6h | 2635.58 | 549.602 | 0.34192 | 0.0654388 |
| Chitin 6h | 2777.74 | 421.291 | 0.385856 | 0.0447583 |
| Epi 6h | 2794.32 | 432.182 | 0.36496 | 0.0825558 |
| SA 6h | 2436.22 | 373.375 | 0.362437 | 0.0209353 |
| Me-JA 6h | 1174.43 | 466.286 | 0.149568 | 0.056603 |
Source Transcript PGSC0003DMT400018519 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G56940.1 | +1 | 0.0 | 654 | 342/409 (84%) | dicarboxylate diiron protein, putative (Crd1) | chr3:21076594-21078269 FORWARD LENGTH=409 |
