Probe CUST_46048_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46048_PI426222305 | JHI_St_60k_v1 | DMT400040582 | GGATGAGACGGCATACAATTAGATACACTCGGAGGGAAGAATTTTAGAGTTGAAAAATTA |
All Microarray Probes Designed to Gene DMG400015688
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46048_PI426222305 | JHI_St_60k_v1 | DMT400040582 | GGATGAGACGGCATACAATTAGATACACTCGGAGGGAAGAATTTTAGAGTTGAAAAATTA |
Microarray Signals from CUST_46048_PI426222305
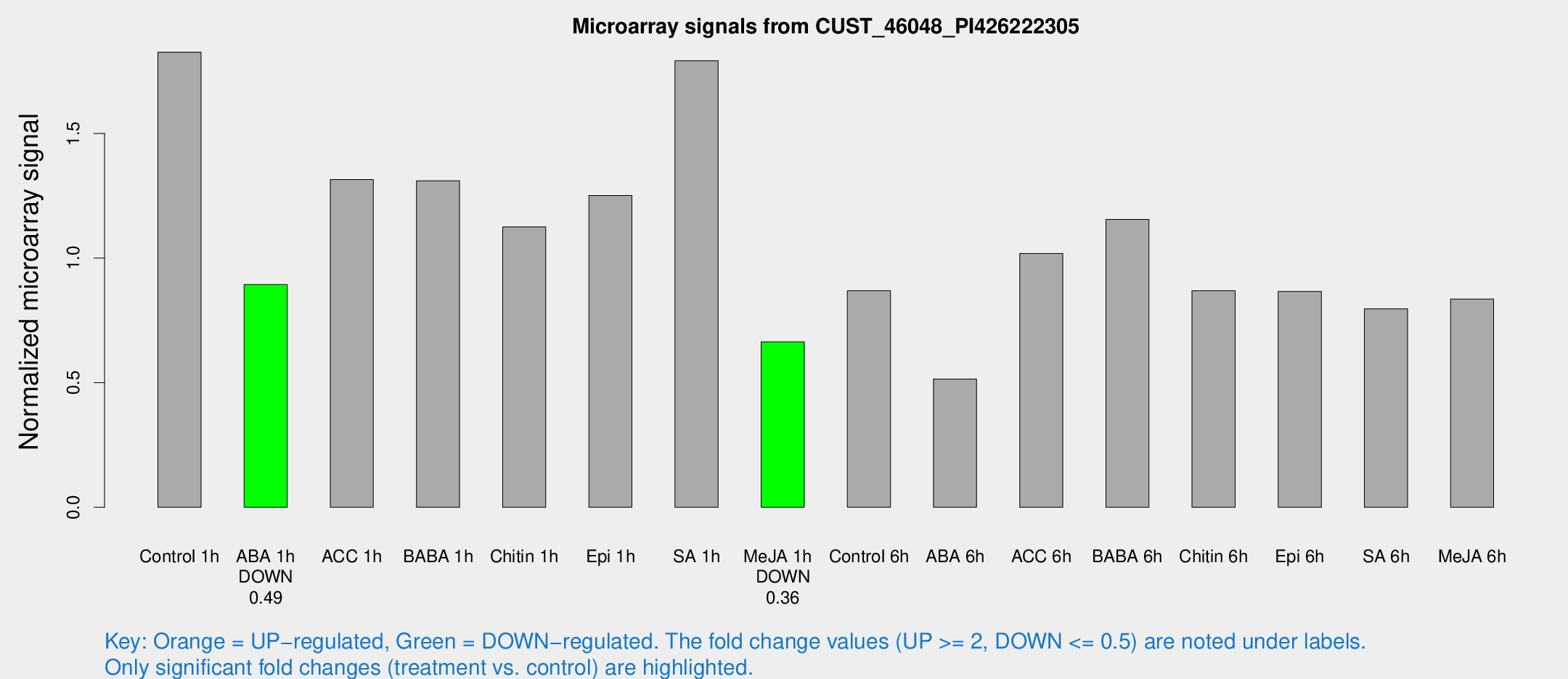
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5106.05 | 547.071 | 1.82613 | 0.105439 |
| ABA 1h | 2203.85 | 199.721 | 0.893701 | 0.0695708 |
| ACC 1h | 4362.19 | 1403.76 | 1.31557 | 0.508797 |
| BABA 1h | 3684.1 | 785.139 | 1.30991 | 0.18353 |
| Chitin 1h | 2803.76 | 162.201 | 1.12565 | 0.0650037 |
| Epi 1h | 3015.78 | 251.457 | 1.25092 | 0.0956506 |
| SA 1h | 5186.9 | 688.744 | 1.79181 | 0.189843 |
| Me-JA 1h | 1510.43 | 127.418 | 0.664266 | 0.0383822 |
| Control 6h | 2531.65 | 511.131 | 0.869407 | 0.120524 |
| ABA 6h | 1567.48 | 306.089 | 0.514797 | 0.0688349 |
| ACC 6h | 3234.54 | 187.075 | 1.01825 | 0.110162 |
| BABA 6h | 3600.38 | 296.467 | 1.15512 | 0.0667029 |
| Chitin 6h | 2561.15 | 148.127 | 0.86892 | 0.0501842 |
| Epi 6h | 2721.67 | 242.514 | 0.866063 | 0.0645306 |
| SA 6h | 2272.66 | 433.161 | 0.796437 | 0.0771692 |
| Me-JA 6h | 2412.75 | 517.03 | 0.835949 | 0.113294 |
Source Transcript PGSC0003DMT400040582 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G38040.1 | +1 | 1e-40 | 152 | 101/338 (30%) | Exostosin family protein | chr4:17867501-17869131 FORWARD LENGTH=425 |
