Probe CUST_46045_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46045_PI426222305 | JHI_St_60k_v1 | DMT400040461 | AAGAAACGGCGTCGTCAAGCGTTCATTGGGAAAACTAATTCAAGACAAATTGCATTGTTT |
All Microarray Probes Designed to Gene DMG400015667
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46045_PI426222305 | JHI_St_60k_v1 | DMT400040461 | AAGAAACGGCGTCGTCAAGCGTTCATTGGGAAAACTAATTCAAGACAAATTGCATTGTTT |
Microarray Signals from CUST_46045_PI426222305
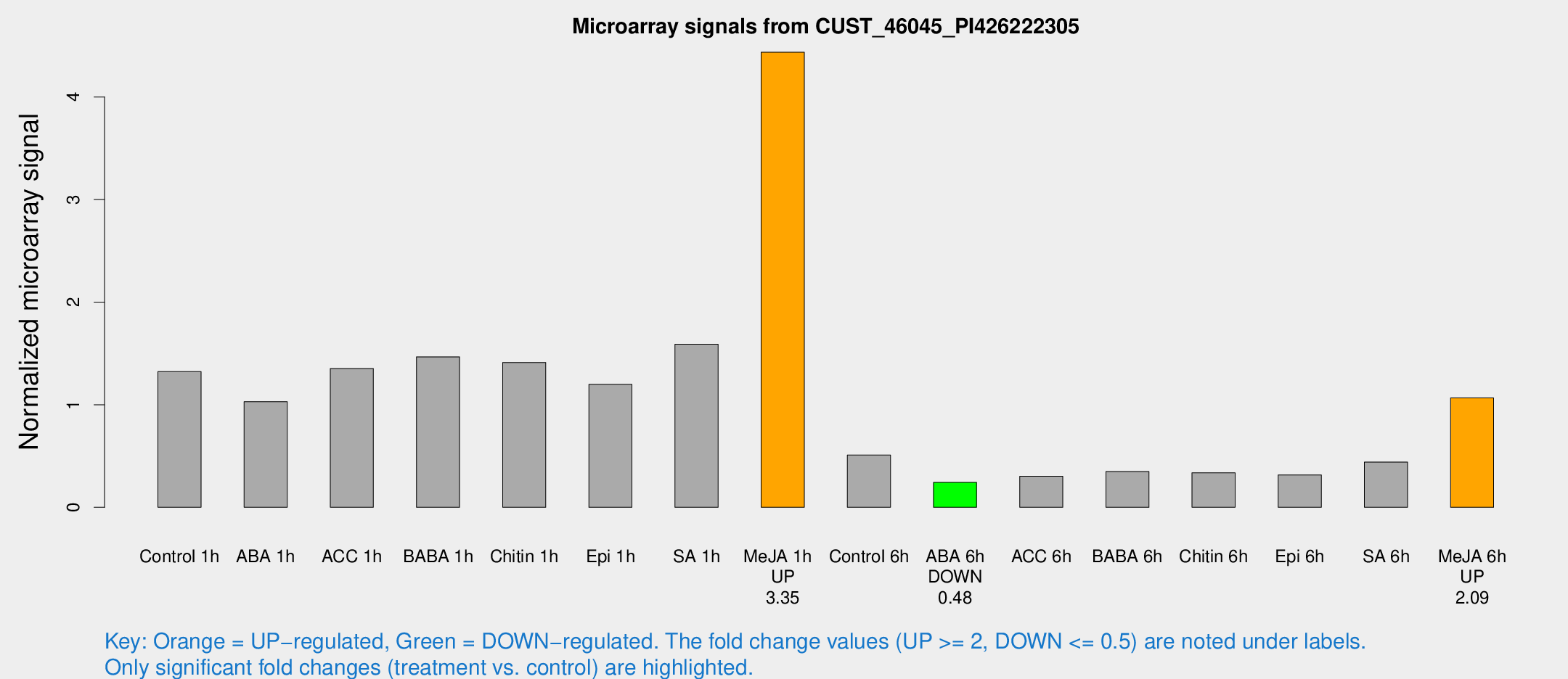
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 26994.4 | 4113.58 | 1.32338 | 0.162589 |
| ABA 1h | 19100.4 | 4236.42 | 1.02998 | 0.162561 |
| ACC 1h | 29526.2 | 7836.16 | 1.35296 | 0.315143 |
| BABA 1h | 29301.4 | 5315.98 | 1.46708 | 0.145148 |
| Chitin 1h | 25415.9 | 1642.36 | 1.41184 | 0.130012 |
| Epi 1h | 20774.9 | 1357.15 | 1.19886 | 0.0893592 |
| SA 1h | 33730 | 6591.19 | 1.59013 | 0.310702 |
| Me-JA 1h | 73659.1 | 9953.35 | 4.43637 | 0.282781 |
| Control 6h | 10512.3 | 1815.12 | 0.509482 | 0.0462028 |
| ABA 6h | 5175.38 | 369.8 | 0.242922 | 0.014026 |
| ACC 6h | 6958.52 | 470.932 | 0.302754 | 0.0375404 |
| BABA 6h | 7802.93 | 451.689 | 0.348709 | 0.0201334 |
| Chitin 6h | 7153.81 | 414.259 | 0.33607 | 0.0241113 |
| Epi 6h | 7471.26 | 1562.24 | 0.315825 | 0.0974182 |
| SA 6h | 9044.99 | 1737.71 | 0.44104 | 0.0450169 |
| Me-JA 6h | 21169 | 1222.57 | 1.0658 | 0.0766697 |
Source Transcript PGSC0003DMT400040461 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
