Probe CUST_45930_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45930_PI426222305 | JHI_St_60k_v1 | DMT400047949 | GTAGTACTAACATTGTTTTATTTGTGTGTATGGGAACAGAGTTCCTTGTGATTACAAAGG |
All Microarray Probes Designed to Gene DMG400018635
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45899_PI426222305 | JHI_St_60k_v1 | DMT400047948 | AATTGTCAGTGTTGATGTTGCTCAGGTTGGATCGTCGAACTGGAACTTCTTGACCCGAAA |
| CUST_45930_PI426222305 | JHI_St_60k_v1 | DMT400047949 | GTAGTACTAACATTGTTTTATTTGTGTGTATGGGAACAGAGTTCCTTGTGATTACAAAGG |
Microarray Signals from CUST_45930_PI426222305
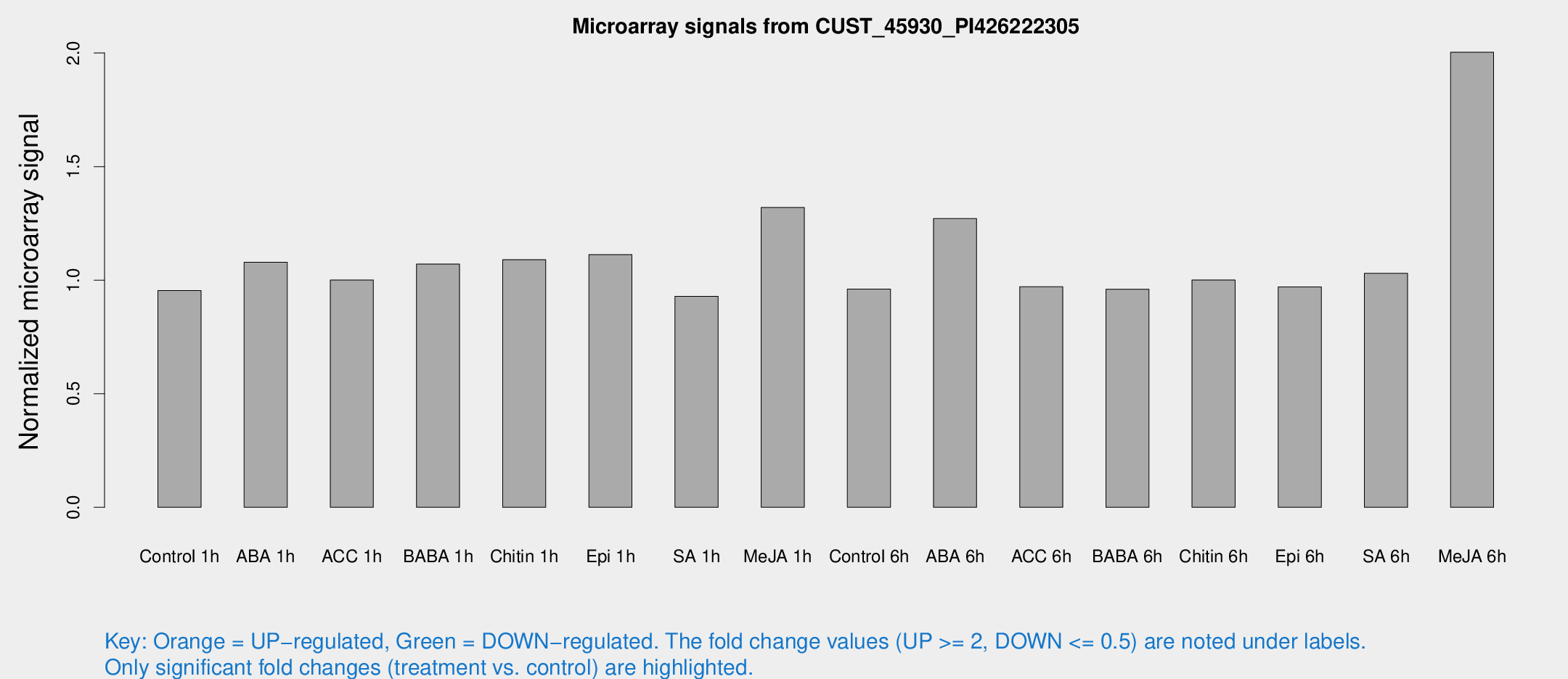
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4.86532 | 2.82275 | 0.954258 | 0.538319 |
| ABA 1h | 4.67651 | 2.70948 | 1.07873 | 0.584288 |
| ACC 1h | 5.40425 | 3.14321 | 1.00058 | 0.579774 |
| BABA 1h | 5.39572 | 2.99531 | 1.07061 | 0.592336 |
| Chitin 1h | 4.84862 | 2.82062 | 1.09012 | 0.594575 |
| Epi 1h | 4.84427 | 2.80992 | 1.11242 | 0.618295 |
| SA 1h | 4.91315 | 2.84542 | 0.928905 | 0.528572 |
| Me-JA 1h | 5.52216 | 2.89527 | 1.3199 | 0.675043 |
| Control 6h | 5.00343 | 2.8988 | 0.961101 | 0.551808 |
| ABA 6h | 7.56596 | 3.06367 | 1.27119 | 0.582069 |
| ACC 6h | 5.95857 | 3.52755 | 0.970812 | 0.562278 |
| BABA 6h | 5.64285 | 3.27407 | 0.960211 | 0.55604 |
| Chitin 6h | 5.58361 | 3.2358 | 1.00082 | 0.579872 |
| Epi 6h | 5.77476 | 3.37337 | 0.969986 | 0.562269 |
| SA 6h | 5.34148 | 3.09859 | 1.02993 | 0.597076 |
| Me-JA 6h | 11.6771 | 3.40643 | 2.00342 | 0.787458 |
Source Transcript PGSC0003DMT400047949 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G45970.1 | +1 | 5e-46 | 130 | 57/81 (70%) | expansin-like A1 | chr3:16896238-16897189 FORWARD LENGTH=265 |
