Probe CUST_45899_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45899_PI426222305 | JHI_St_60k_v1 | DMT400047948 | AATTGTCAGTGTTGATGTTGCTCAGGTTGGATCGTCGAACTGGAACTTCTTGACCCGAAA |
All Microarray Probes Designed to Gene DMG400018635
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45930_PI426222305 | JHI_St_60k_v1 | DMT400047949 | GTAGTACTAACATTGTTTTATTTGTGTGTATGGGAACAGAGTTCCTTGTGATTACAAAGG |
| CUST_45899_PI426222305 | JHI_St_60k_v1 | DMT400047948 | AATTGTCAGTGTTGATGTTGCTCAGGTTGGATCGTCGAACTGGAACTTCTTGACCCGAAA |
Microarray Signals from CUST_45899_PI426222305
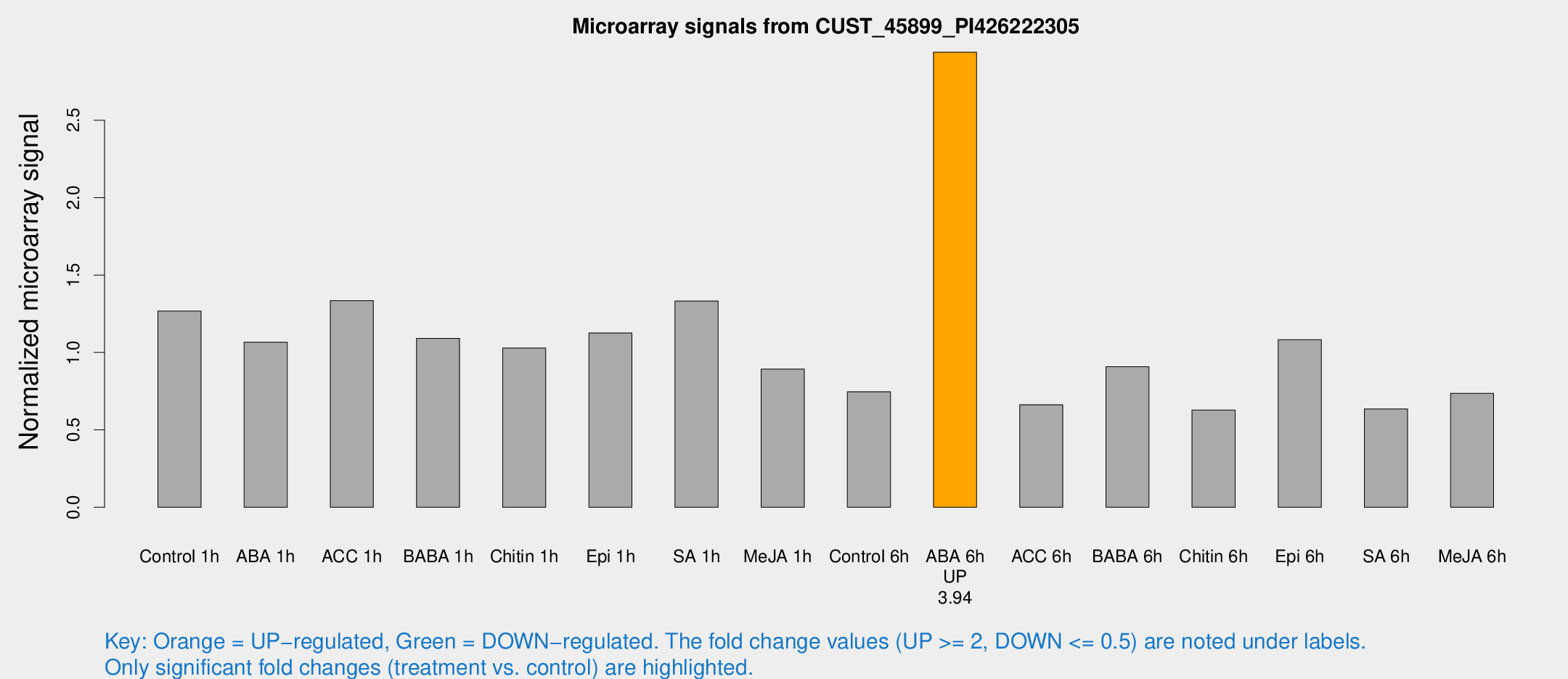
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1439.58 | 425.149 | 1.26727 | 0.279167 |
| ABA 1h | 1001.97 | 112.919 | 1.06631 | 0.102084 |
| ACC 1h | 1470.06 | 212.167 | 1.33408 | 0.101434 |
| BABA 1h | 1114.96 | 118.011 | 1.09129 | 0.124707 |
| Chitin 1h | 987.822 | 139.095 | 1.02818 | 0.152577 |
| Epi 1h | 1024.94 | 59.4396 | 1.12592 | 0.06511 |
| SA 1h | 1461.87 | 194.839 | 1.33183 | 0.102145 |
| Me-JA 1h | 767.284 | 47.6981 | 0.89281 | 0.0974123 |
| Control 6h | 889.906 | 263.554 | 0.746289 | 0.230831 |
| ABA 6h | 3301.75 | 277.527 | 2.93894 | 0.302833 |
| ACC 6h | 846.317 | 216.263 | 0.662302 | 0.0804663 |
| BABA 6h | 1074.03 | 90.4511 | 0.907973 | 0.0919041 |
| Chitin 6h | 719.352 | 120.069 | 0.627945 | 0.105088 |
| Epi 6h | 1349.01 | 322.376 | 1.08315 | 0.254217 |
| SA 6h | 680.472 | 113.008 | 0.635227 | 0.0415052 |
| Me-JA 6h | 822.364 | 195.798 | 0.736149 | 0.140633 |
Source Transcript PGSC0003DMT400047948 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G45970.1 | +1 | 4e-118 | 346 | 165/245 (67%) | expansin-like A1 | chr3:16896238-16897189 FORWARD LENGTH=265 |
