Probe CUST_45767_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45767_PI426222305 | JHI_St_60k_v1 | DMT400055694 | ACTATTTCTTCTTCTAAGTTGTTGGCGAAAAGTCTGAAGAGGGCTCATGACTCGACTTAG |
All Microarray Probes Designed to Gene DMG400021623
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45767_PI426222305 | JHI_St_60k_v1 | DMT400055694 | ACTATTTCTTCTTCTAAGTTGTTGGCGAAAAGTCTGAAGAGGGCTCATGACTCGACTTAG |
Microarray Signals from CUST_45767_PI426222305
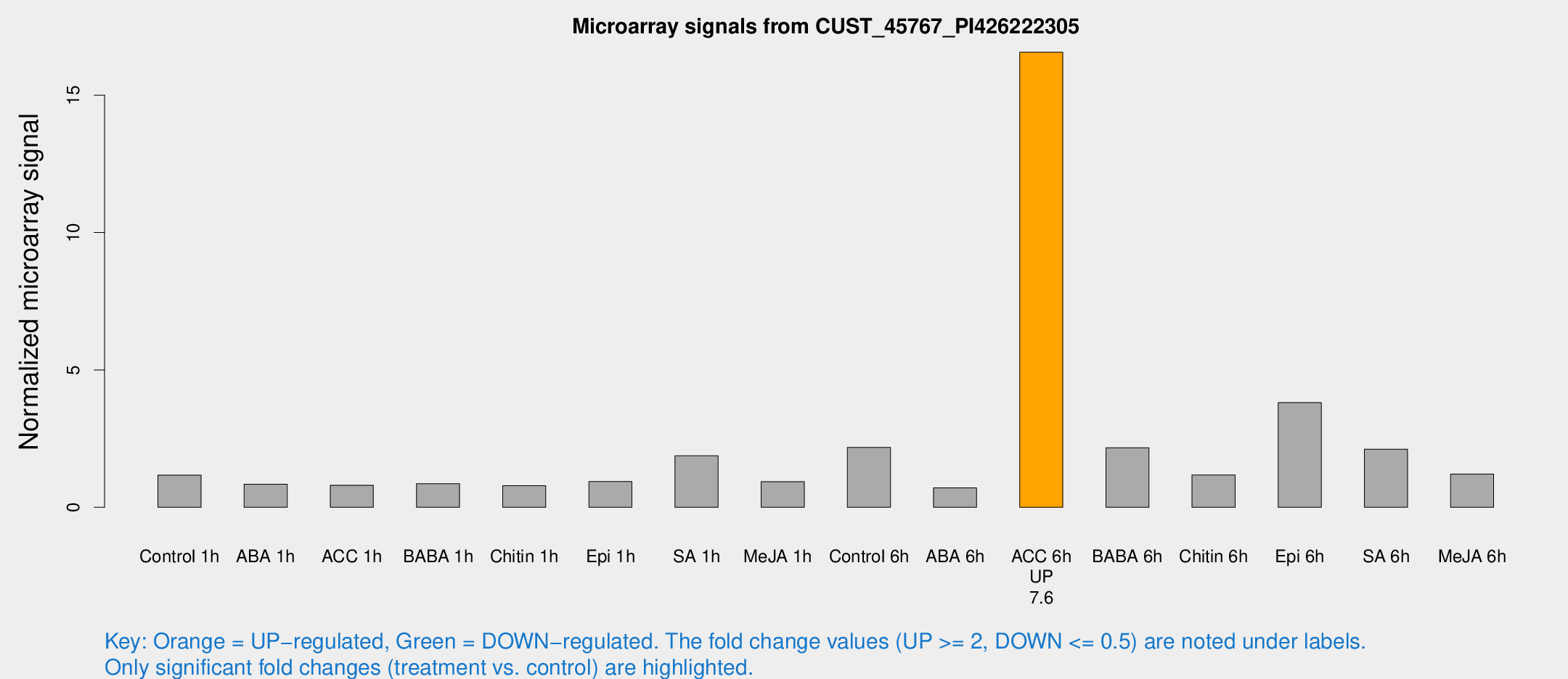
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 11.9514 | 5.84778 | 1.17274 | 0.522793 |
| ABA 1h | 6.15106 | 3.23601 | 0.843284 | 0.446503 |
| ACC 1h | 6.74814 | 3.64648 | 0.803638 | 0.434545 |
| BABA 1h | 6.8526 | 3.53881 | 0.857597 | 0.454076 |
| Chitin 1h | 5.80859 | 3.37808 | 0.787698 | 0.456825 |
| Epi 1h | 6.75921 | 3.35687 | 0.937516 | 0.482722 |
| SA 1h | 16.6111 | 3.49583 | 1.87607 | 0.58012 |
| Me-JA 1h | 6.23863 | 3.43766 | 0.931125 | 0.514513 |
| Control 6h | 21.1403 | 8.29822 | 2.17932 | 0.836934 |
| ABA 6h | 6.16141 | 3.57222 | 0.708116 | 0.410041 |
| ACC 6h | 159.486 | 24.924 | 16.5634 | 2.95177 |
| BABA 6h | 27.8607 | 13.8187 | 2.16944 | 1.71669 |
| Chitin 6h | 10.5131 | 3.82752 | 1.17468 | 0.457285 |
| Epi 6h | 37.5456 | 8.73635 | 3.81285 | 0.829384 |
| SA 6h | 21.6029 | 8.4548 | 2.11719 | 1.60715 |
| Me-JA 6h | 10.3986 | 3.47216 | 1.20892 | 0.465928 |
Source Transcript PGSC0003DMT400055694 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G16740.1 | +1 | 9e-107 | 323 | 196/316 (62%) | Transmembrane amino acid transporter family protein | chr5:5501044-5502856 REVERSE LENGTH=426 |
