Probe CUST_45072_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45072_PI426222305 | JHI_St_60k_v1 | DMT400051115 | CAGAACTTGGAAGGAAAACATTTCAGTGTGATAACTCGTACAAATTCGTGTAATTGCATT |
All Microarray Probes Designed to Gene DMG400019852
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45072_PI426222305 | JHI_St_60k_v1 | DMT400051115 | CAGAACTTGGAAGGAAAACATTTCAGTGTGATAACTCGTACAAATTCGTGTAATTGCATT |
Microarray Signals from CUST_45072_PI426222305
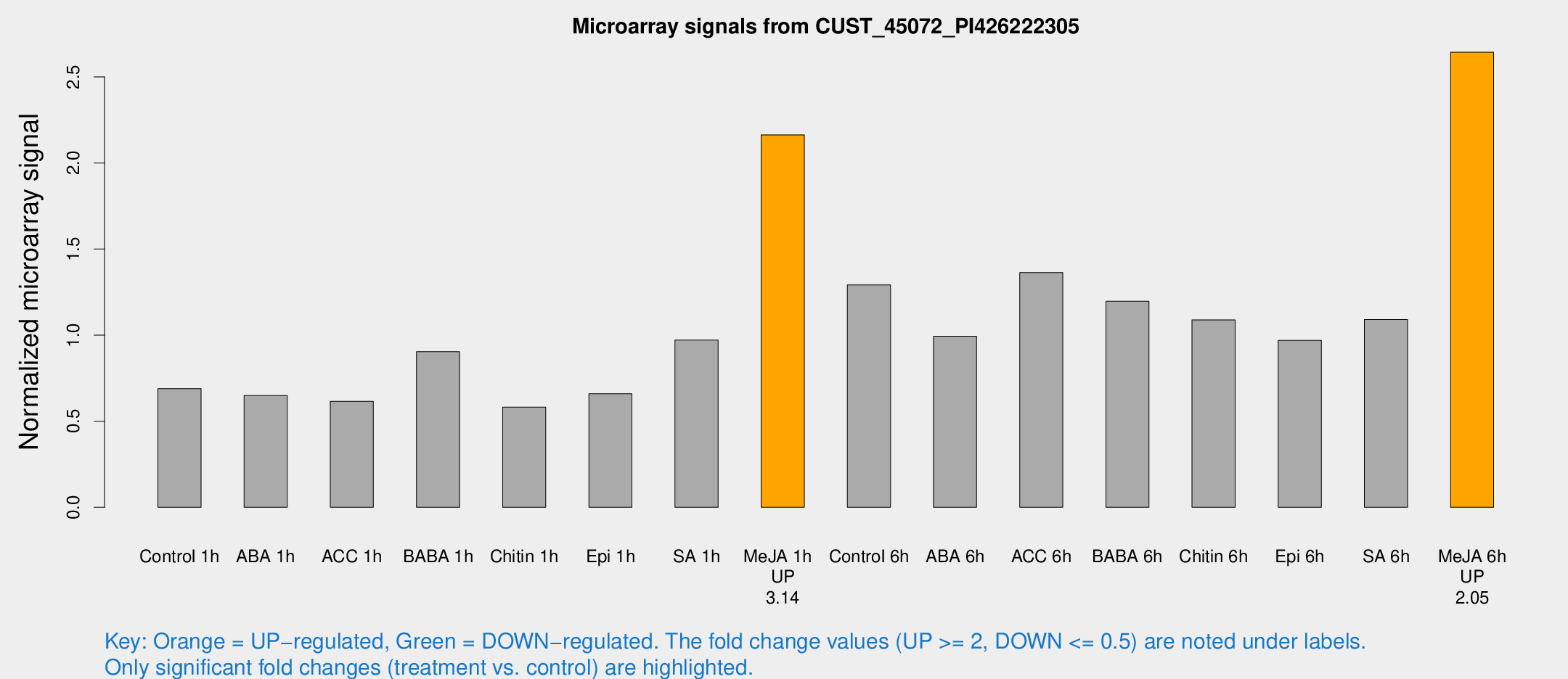
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 602.709 | 81.0591 | 0.689661 | 0.0936619 |
| ABA 1h | 521.657 | 118.101 | 0.649132 | 0.120704 |
| ACC 1h | 632.997 | 238.916 | 0.615721 | 0.229651 |
| BABA 1h | 759.871 | 96.0016 | 0.904044 | 0.0524163 |
| Chitin 1h | 450.966 | 35.0374 | 0.581633 | 0.0338887 |
| Epi 1h | 508.55 | 92.7667 | 0.659504 | 0.129965 |
| SA 1h | 955.606 | 322.314 | 0.971107 | 0.381345 |
| Me-JA 1h | 1540.71 | 208.022 | 2.16335 | 0.125008 |
| Control 6h | 1136.56 | 167.865 | 1.29171 | 0.0811926 |
| ABA 6h | 909.12 | 60.2632 | 0.993235 | 0.0575101 |
| ACC 6h | 1376.09 | 208.697 | 1.36297 | 0.235217 |
| BABA 6h | 1165.53 | 144.711 | 1.19661 | 0.130212 |
| Chitin 6h | 1003.01 | 97.9624 | 1.08818 | 0.139241 |
| Epi 6h | 946.662 | 94.9334 | 0.969387 | 0.176779 |
| SA 6h | 945.406 | 132.131 | 1.09087 | 0.0631736 |
| Me-JA 6h | 2256.82 | 130.38 | 2.64314 | 0.188661 |
Source Transcript PGSC0003DMT400051115 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G14030.1 | +3 | 1e-40 | 146 | 82/150 (55%) | translocon-associated protein beta (TRAPB) family protein | chr5:4526878-4528253 FORWARD LENGTH=195 |
