Probe CUST_45063_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45063_PI426222305 | JHI_St_60k_v1 | DMT400051127 | ATTGGACTAGACATCCCTCATGCTGCCCTTTTGGAAGTTGGAATGGTGTATACAATATAA |
All Microarray Probes Designed to Gene DMG400019859
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45063_PI426222305 | JHI_St_60k_v1 | DMT400051127 | ATTGGACTAGACATCCCTCATGCTGCCCTTTTGGAAGTTGGAATGGTGTATACAATATAA |
Microarray Signals from CUST_45063_PI426222305
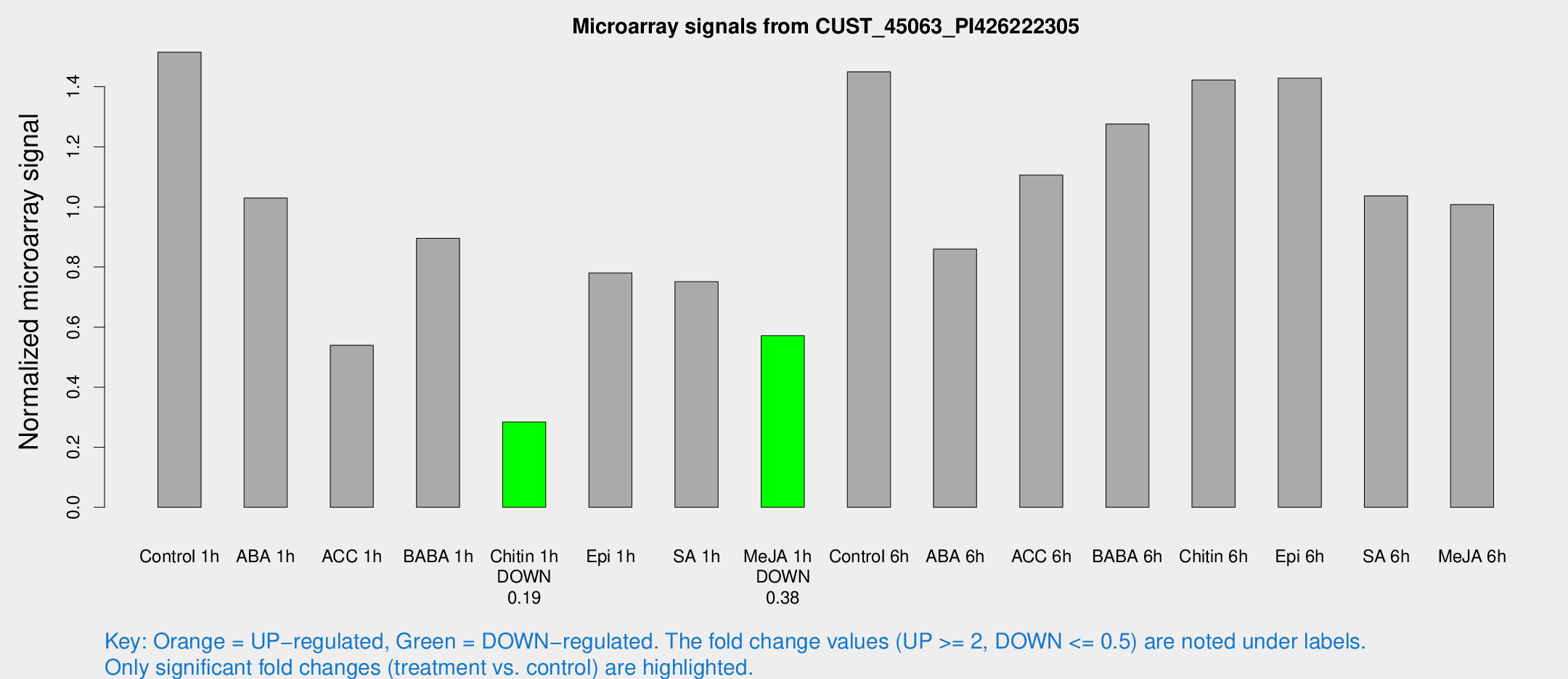
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 39.0429 | 4.22109 | 1.51497 | 0.156604 |
| ABA 1h | 23.8852 | 4.2129 | 1.02963 | 0.258637 |
| ACC 1h | 15.9901 | 5.69504 | 0.53906 | 0.217575 |
| BABA 1h | 23.964 | 7.1675 | 0.895711 | 0.201964 |
| Chitin 1h | 6.5007 | 3.27291 | 0.283803 | 0.142691 |
| Epi 1h | 17.2316 | 3.34614 | 0.780398 | 0.151569 |
| SA 1h | 23.0231 | 7.45705 | 0.75123 | 0.325708 |
| Me-JA 1h | 11.8742 | 3.47346 | 0.571117 | 0.16717 |
| Control 6h | 41.6465 | 14.9358 | 1.44987 | 0.454798 |
| ABA 6h | 24.3051 | 4.68366 | 0.859618 | 0.191618 |
| ACC 6h | 32.6064 | 4.50738 | 1.10604 | 0.157557 |
| BABA 6h | 39.1716 | 10.9702 | 1.27611 | 0.374494 |
| Chitin 6h | 40.9567 | 10.4944 | 1.42266 | 0.320961 |
| Epi 6h | 41.4672 | 4.72787 | 1.42876 | 0.166982 |
| SA 6h | 29.6586 | 10.9638 | 1.03698 | 0.303323 |
| Me-JA 6h | 28.9841 | 9.29048 | 1.0076 | 0.343689 |
Source Transcript PGSC0003DMT400051127 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G40540.1 | +1 | 0.0 | 658 | 380/801 (47%) | potassium transporter 2 | chr2:16931445-16934516 FORWARD LENGTH=794 |
