Probe CUST_45060_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45060_PI426222305 | JHI_St_60k_v1 | DMT400017248 | GGTCAGCTGCTAGAAAAAGGGTATGTCATCACGTTAAGGAATGATACATGTGAAATTTCT |
All Microarray Probes Designed to Gene DMG400006724
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45060_PI426222305 | JHI_St_60k_v1 | DMT400017248 | GGTCAGCTGCTAGAAAAAGGGTATGTCATCACGTTAAGGAATGATACATGTGAAATTTCT |
Microarray Signals from CUST_45060_PI426222305
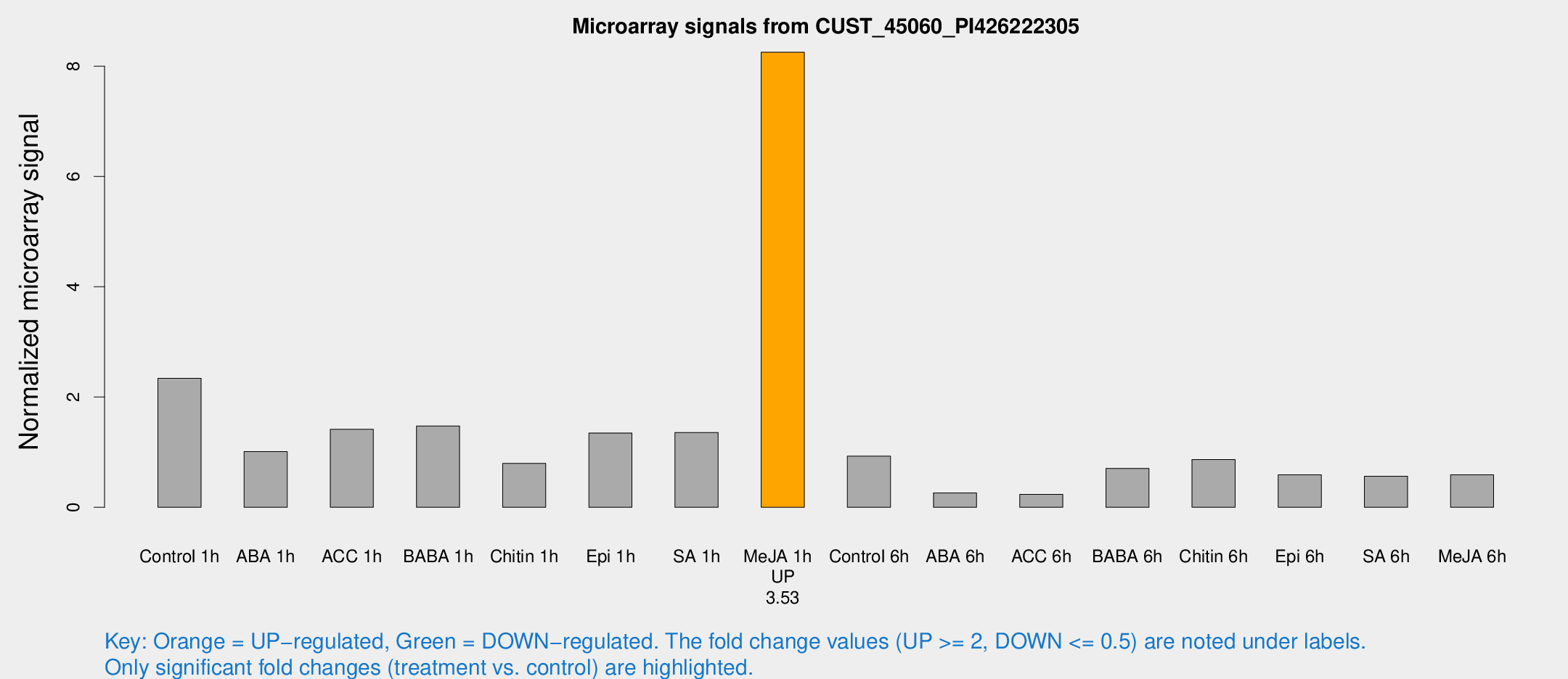
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 158.378 | 9.87202 | 2.33932 | 0.145562 |
| ABA 1h | 82.6828 | 43.261 | 1.01003 | 0.636523 |
| ACC 1h | 108.519 | 35.8344 | 1.41395 | 0.404307 |
| BABA 1h | 96.8988 | 8.99203 | 1.47535 | 0.104584 |
| Chitin 1h | 52.0676 | 13.0446 | 0.796789 | 0.297388 |
| Epi 1h | 83.7691 | 19.1167 | 1.34751 | 0.335815 |
| SA 1h | 100.865 | 27.3143 | 1.35767 | 0.401049 |
| Me-JA 1h | 466.57 | 72.7894 | 8.25423 | 1.20274 |
| Control 6h | 66.498 | 14.4401 | 0.927258 | 0.14747 |
| ABA 6h | 22.4545 | 7.60946 | 0.261863 | 0.146908 |
| ACC 6h | 19.8549 | 6.33635 | 0.232981 | 0.088059 |
| BABA 6h | 56.3838 | 12.7334 | 0.705188 | 0.155789 |
| Chitin 6h | 62.7006 | 5.80059 | 0.865284 | 0.100004 |
| Epi 6h | 48.3077 | 11.4784 | 0.590185 | 0.225308 |
| SA 6h | 41.1466 | 12.8795 | 0.563628 | 0.137225 |
| Me-JA 6h | 45.3796 | 16.4808 | 0.590506 | 0.177449 |
Source Transcript PGSC0003DMT400017248 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
