Probe CUST_44938_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_44938_PI426222305 | JHI_St_60k_v1 | DMT400058685 | GCTGAAGGCCACACAGGAATGCTGATATATTCTTTATATATATTAATGCTGCTAACTTCA |
All Microarray Probes Designed to Gene DMG403022797
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_44938_PI426222305 | JHI_St_60k_v1 | DMT400058685 | GCTGAAGGCCACACAGGAATGCTGATATATTCTTTATATATATTAATGCTGCTAACTTCA |
Microarray Signals from CUST_44938_PI426222305
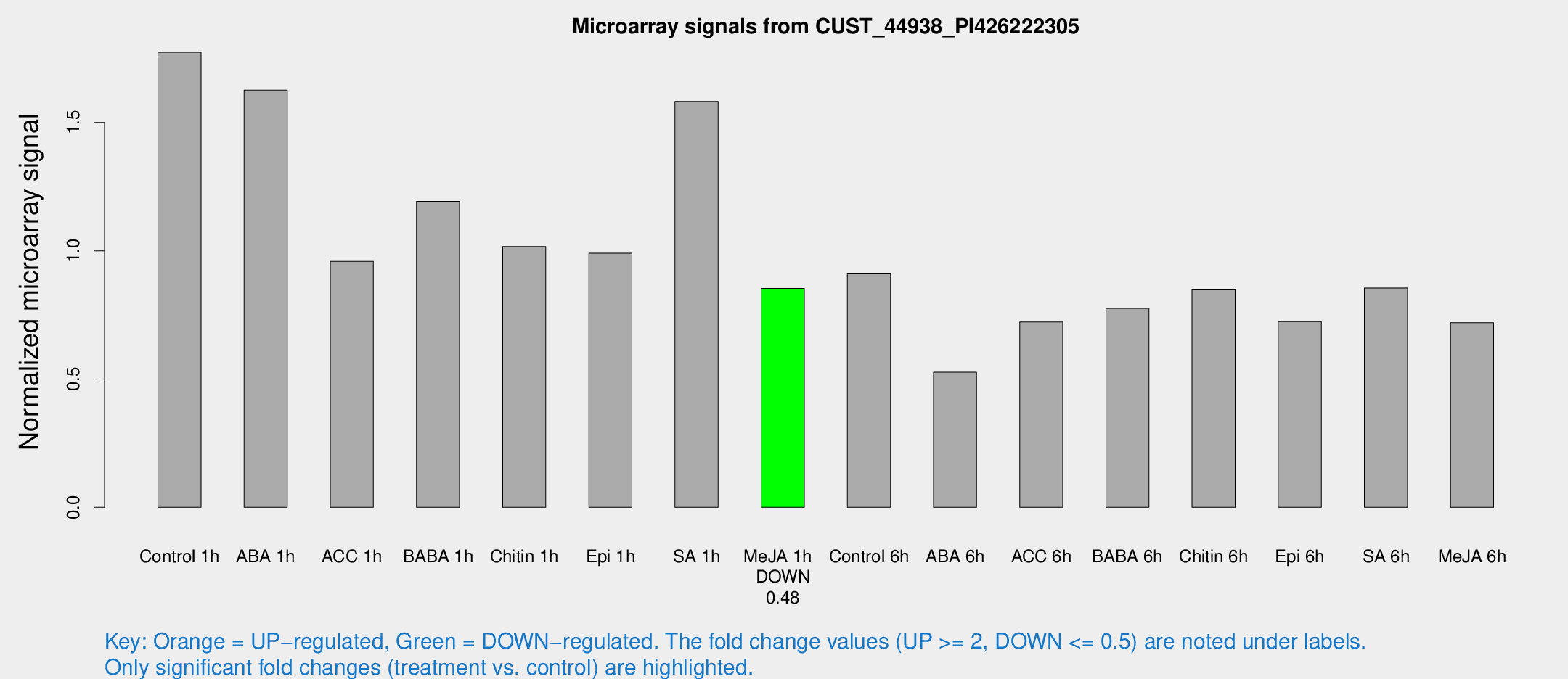
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 54.1685 | 7.61478 | 1.7737 | 0.163468 |
| ABA 1h | 43.2901 | 4.42539 | 1.62582 | 0.167451 |
| ACC 1h | 29.8976 | 4.47461 | 0.95879 | 0.152464 |
| BABA 1h | 35.521 | 6.50127 | 1.193 | 0.159611 |
| Chitin 1h | 27.524 | 4.0979 | 1.01647 | 0.166515 |
| Epi 1h | 27.8651 | 7.34339 | 0.990319 | 0.309123 |
| SA 1h | 52.1768 | 14.5768 | 1.58207 | 0.347979 |
| Me-JA 1h | 21.1798 | 3.83875 | 0.853096 | 0.156808 |
| Control 6h | 32.2704 | 11.7755 | 0.909771 | 0.353198 |
| ABA 6h | 20.2384 | 7.13561 | 0.526585 | 0.261785 |
| ACC 6h | 25.4319 | 5.0195 | 0.722549 | 0.189667 |
| BABA 6h | 27.3367 | 5.98659 | 0.775878 | 0.150923 |
| Chitin 6h | 27.8975 | 4.68973 | 0.847826 | 0.166141 |
| Epi 6h | 25.5027 | 5.00222 | 0.72379 | 0.207336 |
| SA 6h | 35.2664 | 15.3757 | 0.855094 | 0.916516 |
| Me-JA 6h | 25.8947 | 9.22687 | 0.719461 | 0.32081 |
Source Transcript PGSC0003DMT400058685 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G14820.3 | +2 | 2e-65 | 218 | 107/162 (66%) | Sec14p-like phosphatidylinositol transfer family protein | chr1:5105237-5106793 REVERSE LENGTH=252 |
