Probe CUST_44891_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_44891_PI426222305 | JHI_St_60k_v1 | DMT400051416 | ATATATGTGAATTCTGCGTTTGAAAGCTCTACTGGTTATCGCGCTGACGAAGTCCTCGGT |
All Microarray Probes Designed to Gene DMG400019971
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_44891_PI426222305 | JHI_St_60k_v1 | DMT400051416 | ATATATGTGAATTCTGCGTTTGAAAGCTCTACTGGTTATCGCGCTGACGAAGTCCTCGGT |
Microarray Signals from CUST_44891_PI426222305
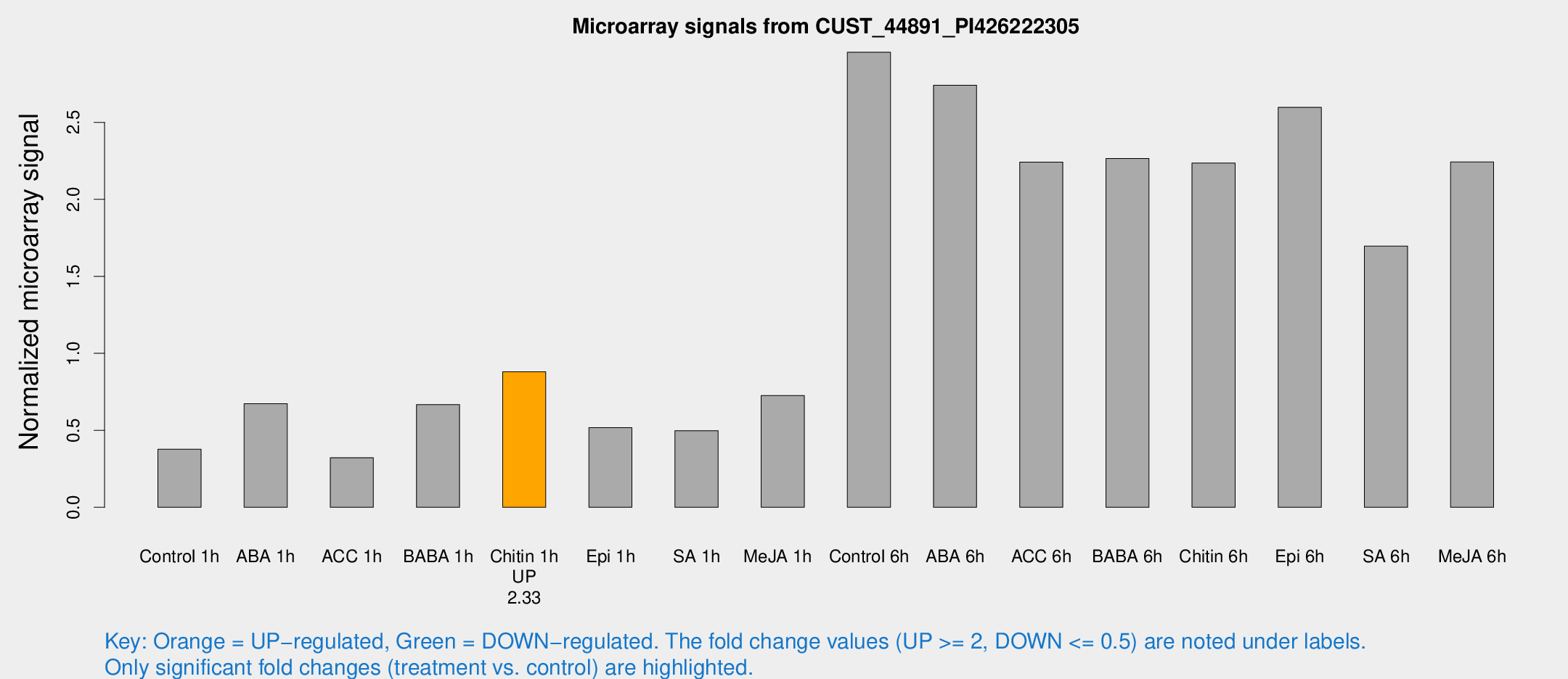
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 142.652 | 12.5637 | 0.377468 | 0.0237344 |
| ABA 1h | 225.266 | 22.6408 | 0.672578 | 0.066721 |
| ACC 1h | 124.749 | 9.32769 | 0.322134 | 0.0224865 |
| BABA 1h | 251.862 | 50.6574 | 0.667033 | 0.081104 |
| Chitin 1h | 298.057 | 19.2227 | 0.880512 | 0.113655 |
| Epi 1h | 168.286 | 10.3161 | 0.517366 | 0.0316317 |
| SA 1h | 198.131 | 35.3175 | 0.497716 | 0.0612405 |
| Me-JA 1h | 231.674 | 48.8546 | 0.726384 | 0.0875445 |
| Control 6h | 1189.65 | 276.426 | 2.95553 | 0.555888 |
| ABA 6h | 1145.43 | 226.566 | 2.7414 | 0.506477 |
| ACC 6h | 993.178 | 172.535 | 2.2423 | 0.129808 |
| BABA 6h | 994.014 | 194.974 | 2.26499 | 0.430778 |
| Chitin 6h | 916.559 | 142.707 | 2.23532 | 0.37569 |
| Epi 6h | 1178.62 | 326.132 | 2.59752 | 0.769673 |
| SA 6h | 680.008 | 190.024 | 1.69643 | 0.539523 |
| Me-JA 6h | 929.941 | 263.722 | 2.2429 | 0.618715 |
Source Transcript PGSC0003DMT400051416 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G68050.1 | +3 | 0.0 | 958 | 475/582 (82%) | flavin-binding, kelch repeat, f box 1 | chr1:25508737-25510697 FORWARD LENGTH=619 |
