Probe CUST_44888_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_44888_PI426222305 | JHI_St_60k_v1 | DMT400038861 | GTACGAAACAACTCTGTAATTAACAGTCTCAATTTTCCGAGGCATTTGACATATTCAGTA |
All Microarray Probes Designed to Gene DMG400015019
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_44888_PI426222305 | JHI_St_60k_v1 | DMT400038861 | GTACGAAACAACTCTGTAATTAACAGTCTCAATTTTCCGAGGCATTTGACATATTCAGTA |
Microarray Signals from CUST_44888_PI426222305
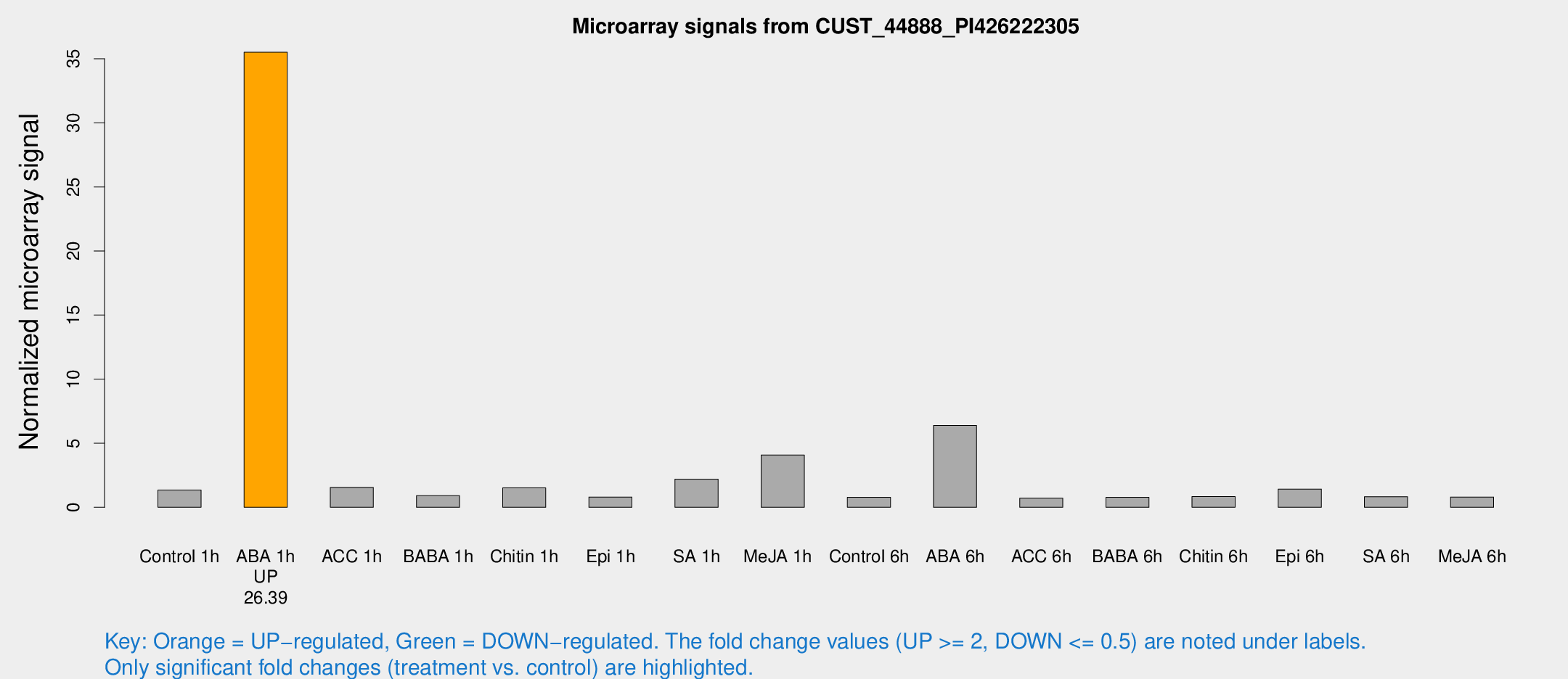
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 13.4674 | 5.42906 | 1.3457 | 0.465214 |
| ABA 1h | 275.396 | 32.5144 | 35.5078 | 2.24565 |
| ACC 1h | 15.794 | 5.64391 | 1.55192 | 0.509002 |
| BABA 1h | 7.94072 | 3.63006 | 0.908776 | 0.453581 |
| Chitin 1h | 11.9902 | 3.43442 | 1.51646 | 0.459531 |
| Epi 1h | 6.00667 | 3.35188 | 0.802898 | 0.446498 |
| SA 1h | 19.8443 | 3.56088 | 2.19772 | 0.409247 |
| Me-JA 1h | 28.9343 | 3.89119 | 4.0857 | 0.630989 |
| Control 6h | 6.88926 | 3.57171 | 0.787433 | 0.417073 |
| ABA 6h | 62.761 | 14.9389 | 6.38109 | 1.47752 |
| ACC 6h | 7.26698 | 4.30846 | 0.717262 | 0.415387 |
| BABA 6h | 7.66669 | 3.98103 | 0.782437 | 0.41435 |
| Chitin 6h | 7.81911 | 3.98844 | 0.844427 | 0.438053 |
| Epi 6h | 14.7357 | 4.13182 | 1.42579 | 0.474145 |
| SA 6h | 7.21404 | 3.70476 | 0.830205 | 0.438299 |
| Me-JA 6h | 7.07791 | 3.52739 | 0.803197 | 0.41685 |
Source Transcript PGSC0003DMT400038861 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G55090.1 | +1 | 2e-104 | 318 | 169/370 (46%) | mitogen-activated protein kinase kinase kinase 15 | chr5:22356852-22358198 REVERSE LENGTH=448 |
