Probe CUST_4478_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_4478_PI426222305 | JHI_St_60k_v1 | DMT400020916 | CAACGTGGAGCGATCGTTACAGTTATGGATTCGATAGAATTCAATAACTTTATATGTCTC |
All Microarray Probes Designed to Gene DMG400008098
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_4478_PI426222305 | JHI_St_60k_v1 | DMT400020916 | CAACGTGGAGCGATCGTTACAGTTATGGATTCGATAGAATTCAATAACTTTATATGTCTC |
Microarray Signals from CUST_4478_PI426222305
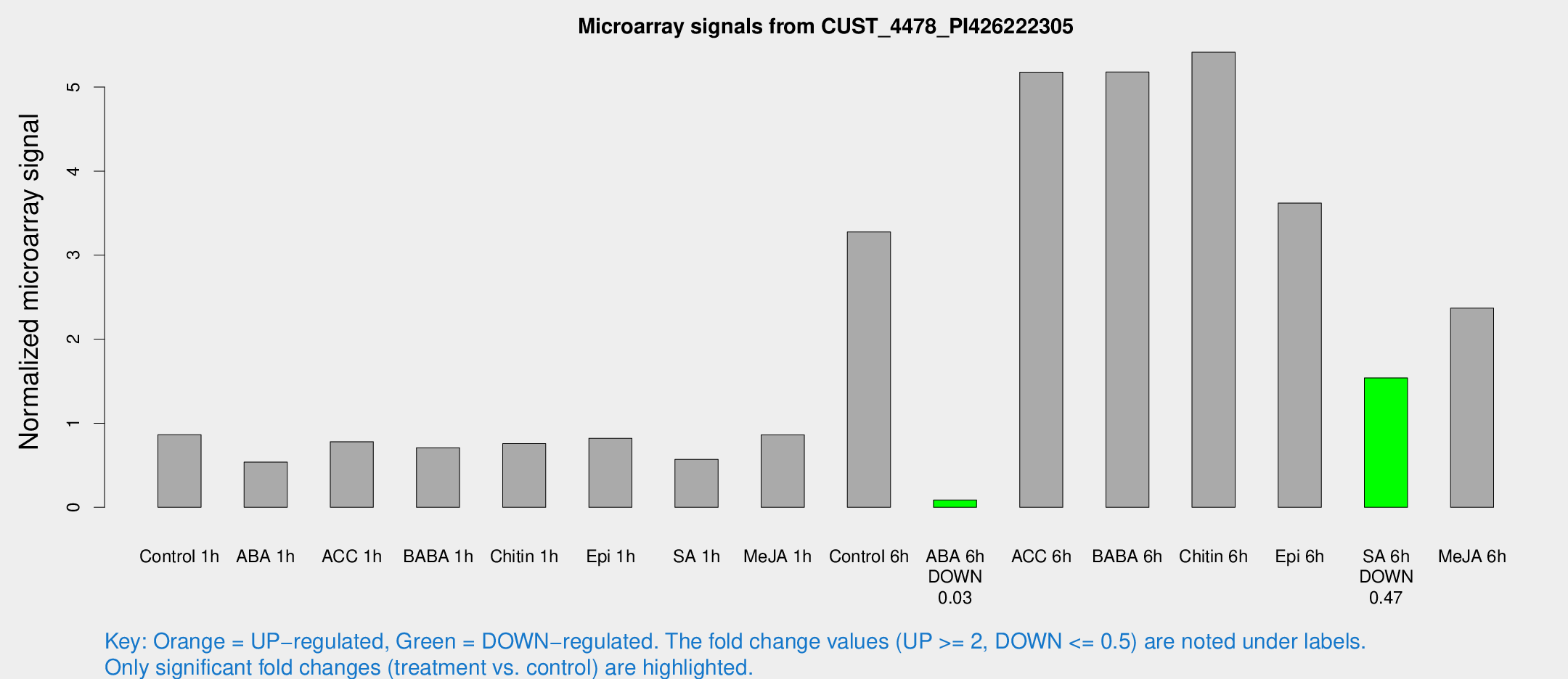
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 104.518 | 14.8862 | 0.863559 | 0.0848494 |
| ABA 1h | 58.9682 | 11.743 | 0.537142 | 0.0929923 |
| ACC 1h | 100.888 | 26.2519 | 0.780496 | 0.155533 |
| BABA 1h | 88.7736 | 25.6006 | 0.709514 | 0.160953 |
| Chitin 1h | 85.6518 | 21.5679 | 0.757349 | 0.227851 |
| Epi 1h | 86.2602 | 13.307 | 0.820302 | 0.123095 |
| SA 1h | 70.6359 | 9.68913 | 0.570341 | 0.0429306 |
| Me-JA 1h | 83.7485 | 5.97021 | 0.862715 | 0.0597451 |
| Control 6h | 419.565 | 102.663 | 3.2765 | 0.664835 |
| ABA 6h | 13.4828 | 6.29829 | 0.0870001 | 0.0509632 |
| ACC 6h | 826.877 | 349.805 | 5.177 | 1.39564 |
| BABA 6h | 858.173 | 391.92 | 5.17855 | 2.6689 |
| Chitin 6h | 700.007 | 107.077 | 5.41454 | 0.75181 |
| Epi 6h | 490.889 | 61.3391 | 3.61989 | 0.76514 |
| SA 6h | 180.156 | 10.9481 | 1.5394 | 0.187711 |
| Me-JA 6h | 340.847 | 119.179 | 2.37004 | 1.14115 |
Source Transcript PGSC0003DMT400020916 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
