Probe CUST_44370_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_44370_PI426222305 | JHI_St_60k_v1 | DMT400030976 | CGATGAAATTTTGAGCAAACGGAAGGAATTTCTTAGCCCTTCTATGTTTTACTTCTATGA |
All Microarray Probes Designed to Gene DMG400011868
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_44370_PI426222305 | JHI_St_60k_v1 | DMT400030976 | CGATGAAATTTTGAGCAAACGGAAGGAATTTCTTAGCCCTTCTATGTTTTACTTCTATGA |
Microarray Signals from CUST_44370_PI426222305
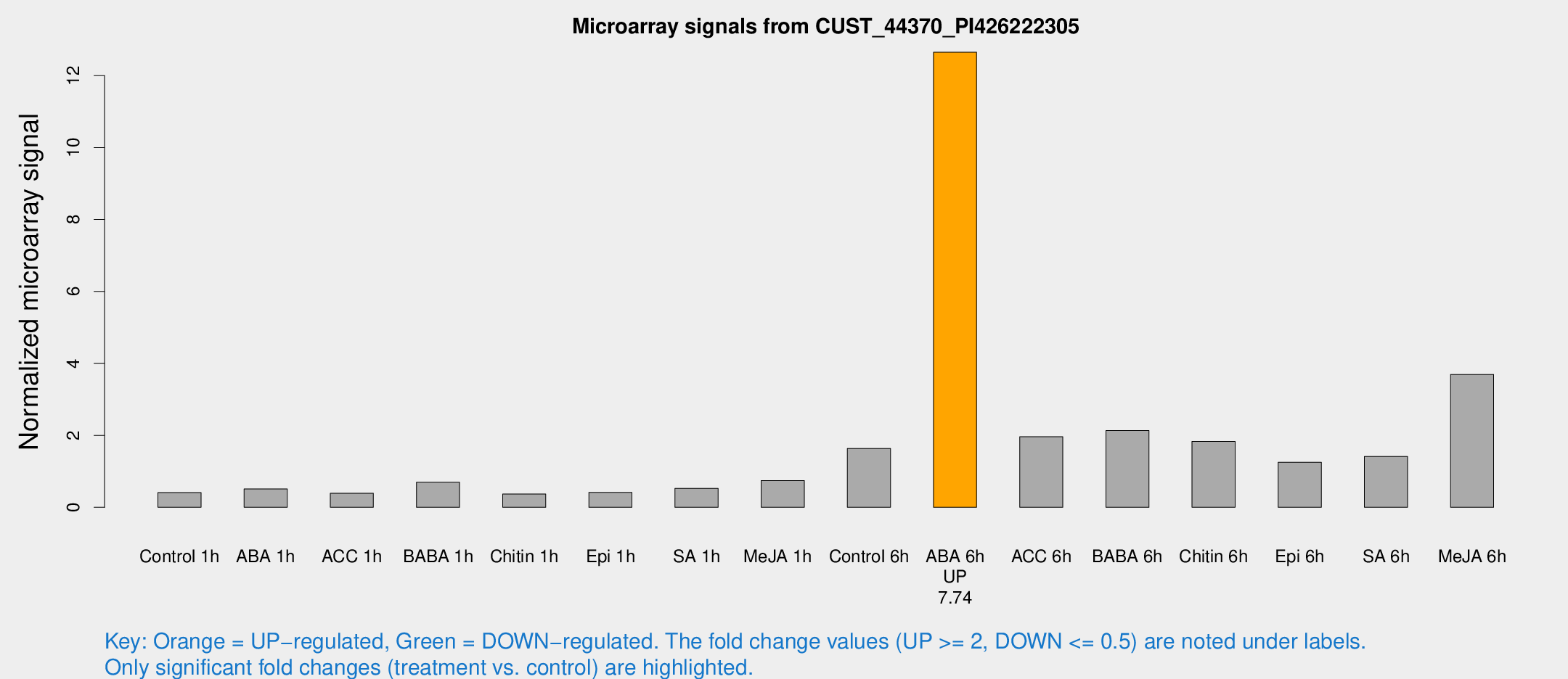
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8.76223 | 3.69825 | 0.409046 | 0.188879 |
| ABA 1h | 8.8036 | 2.97138 | 0.510514 | 0.20008 |
| ACC 1h | 7.48942 | 3.28767 | 0.390002 | 0.179133 |
| BABA 1h | 13.1177 | 3.28536 | 0.699804 | 0.194508 |
| Chitin 1h | 6.12822 | 3.07703 | 0.367506 | 0.186801 |
| Epi 1h | 6.63794 | 3.03357 | 0.41419 | 0.192808 |
| SA 1h | 9.99609 | 3.07842 | 0.526642 | 0.165184 |
| Me-JA 1h | 12.2603 | 3.43468 | 0.742887 | 0.326418 |
| Control 6h | 33.6267 | 11.7726 | 1.63544 | 0.521643 |
| ABA 6h | 257.691 | 53.1702 | 12.6507 | 1.91126 |
| ACC 6h | 43.328 | 9.63457 | 1.96127 | 0.210044 |
| BABA 6h | 49.6431 | 18.3073 | 2.13239 | 0.763393 |
| Chitin 6h | 35.9766 | 4.07598 | 1.83007 | 0.209439 |
| Epi 6h | 26.8163 | 4.48485 | 1.25364 | 0.323157 |
| SA 6h | 38.8731 | 16.948 | 1.41471 | 2.22993 |
| Me-JA 6h | 76.4838 | 24.192 | 3.6945 | 1.12177 |
Source Transcript PGSC0003DMT400030976 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G08860.1 | +2 | 0.0 | 687 | 344/439 (78%) | PYRIMIDINE 4 | chr3:2696754-2699087 REVERSE LENGTH=481 |
