Probe CUST_4421_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_4421_PI426222305 | JHI_St_60k_v1 | DMT400070430 | TTGTTTTTCTTTTGCACGGACCGTTCTGGACGTGAATGCACTTGCTGGATCTTTTGGCAT |
All Microarray Probes Designed to Gene DMG400027380
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_4421_PI426222305 | JHI_St_60k_v1 | DMT400070430 | TTGTTTTTCTTTTGCACGGACCGTTCTGGACGTGAATGCACTTGCTGGATCTTTTGGCAT |
Microarray Signals from CUST_4421_PI426222305
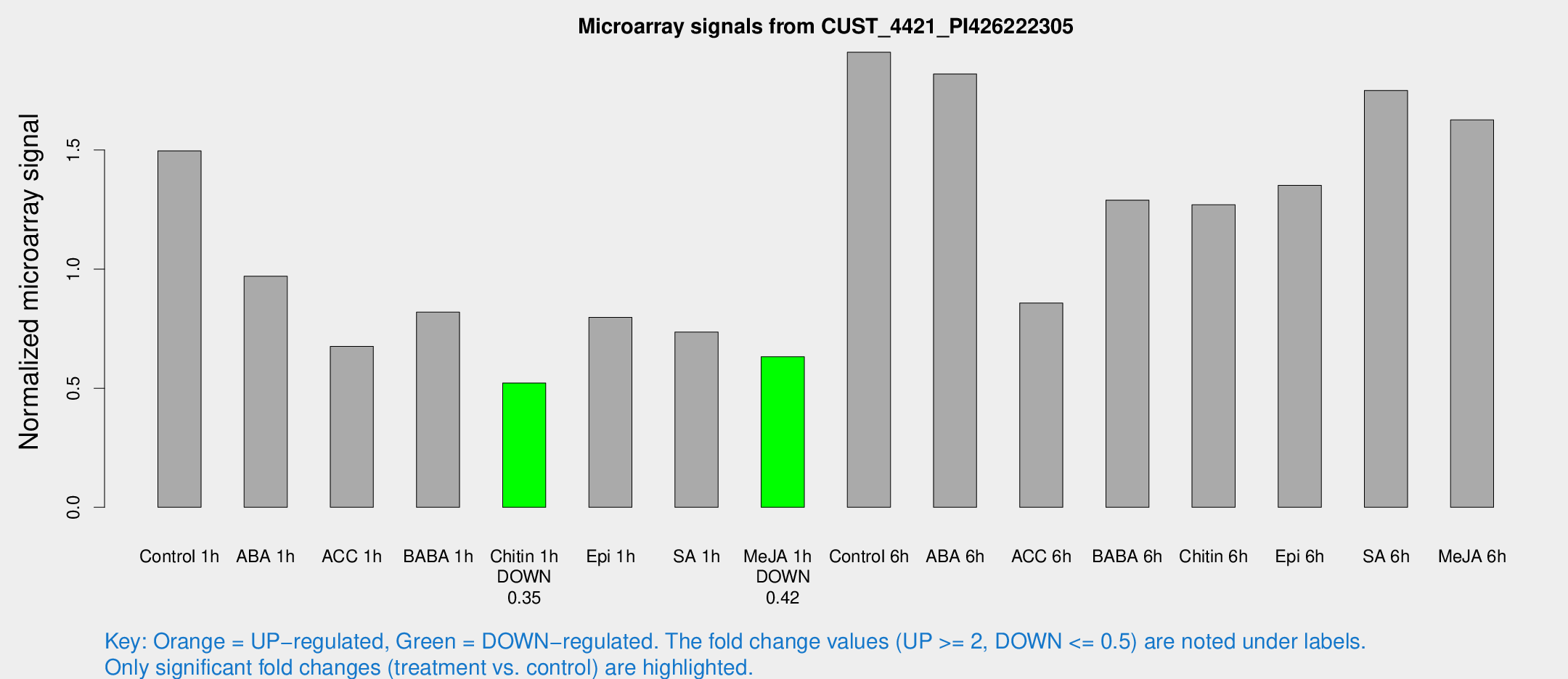
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 17.646 | 3.39522 | 1.49619 | 0.288765 |
| ABA 1h | 10.6269 | 3.10422 | 0.969992 | 0.324103 |
| ACC 1h | 8.95492 | 3.50085 | 0.67559 | 0.315422 |
| BABA 1h | 10.556 | 3.87967 | 0.819371 | 0.349195 |
| Chitin 1h | 5.50229 | 3.19841 | 0.521601 | 0.299973 |
| Epi 1h | 8.65361 | 3.22069 | 0.797583 | 0.34417 |
| SA 1h | 9.46336 | 3.21819 | 0.735558 | 0.289342 |
| Me-JA 1h | 6.11842 | 3.28202 | 0.632159 | 0.341142 |
| Control 6h | 26.6105 | 9.0905 | 1.91075 | 0.739853 |
| ABA 6h | 26.4973 | 8.43664 | 1.81936 | 0.779083 |
| ACC 6h | 11.7774 | 4.08668 | 0.857727 | 0.322542 |
| BABA 6h | 22.0266 | 9.88442 | 1.28975 | 0.816268 |
| Chitin 6h | 16.286 | 3.77914 | 1.27018 | 0.31024 |
| Epi 6h | 20.9835 | 6.83922 | 1.3516 | 0.801264 |
| SA 6h | 22.7772 | 7.77412 | 1.75015 | 0.906885 |
| Me-JA 6h | 22.676 | 9.18244 | 1.6265 | 0.596585 |
Source Transcript PGSC0003DMT400070430 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G51700.1 | +1 | 7e-84 | 274 | 127/210 (60%) | protein binding;zinc ion binding | chr5:21001373-21003143 REVERSE LENGTH=226 |
