Probe CUST_44105_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_44105_PI426222305 | JHI_St_60k_v1 | DMT400055166 | CTGTGTGTATAATGTTGGCTTTGGTTTCTCGGTTGGTTACTCAACTAATATAGTTATTAG |
All Microarray Probes Designed to Gene DMG400021411
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_44105_PI426222305 | JHI_St_60k_v1 | DMT400055166 | CTGTGTGTATAATGTTGGCTTTGGTTTCTCGGTTGGTTACTCAACTAATATAGTTATTAG |
Microarray Signals from CUST_44105_PI426222305
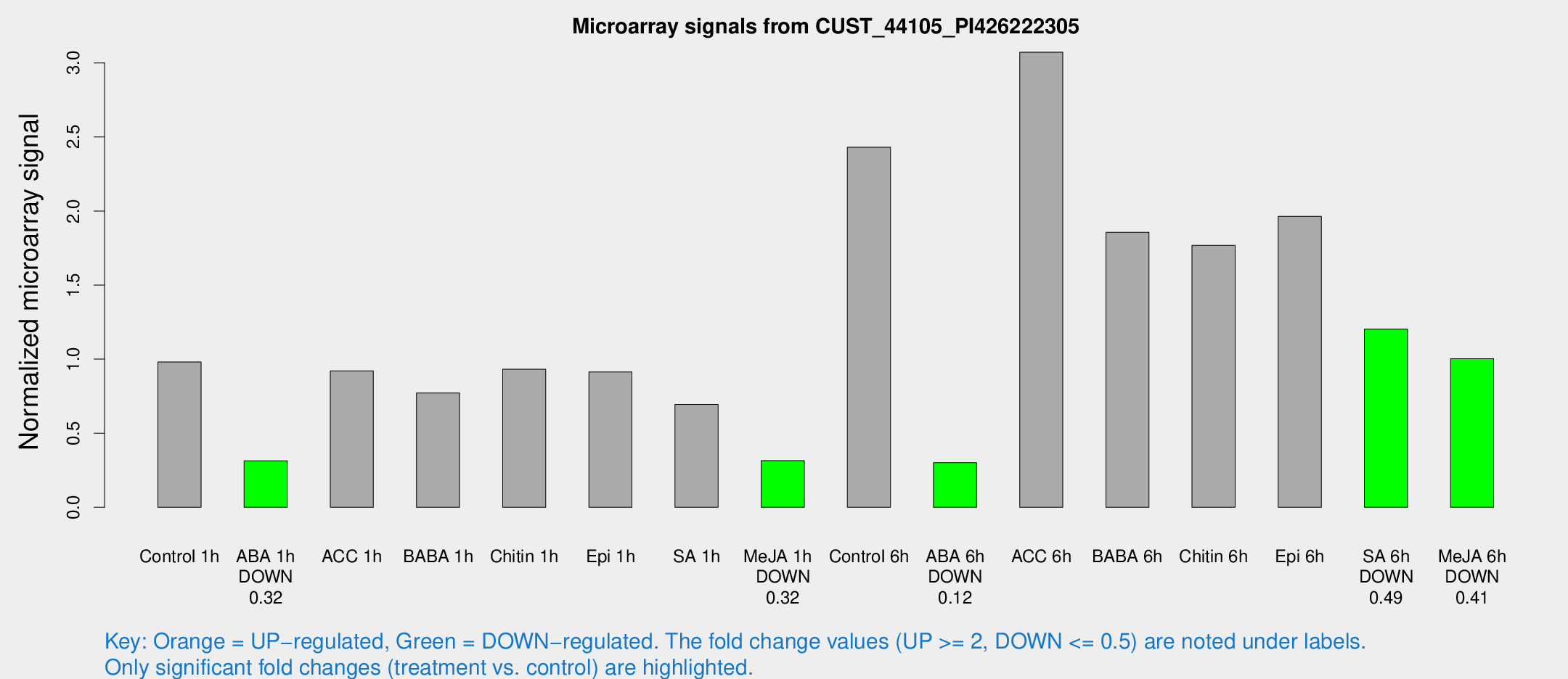
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 357.459 | 25.0179 | 0.980667 | 0.057334 |
| ABA 1h | 105.459 | 23.251 | 0.314383 | 0.0882506 |
| ACC 1h | 360.578 | 73.4637 | 0.921589 | 0.140798 |
| BABA 1h | 278.093 | 45.5431 | 0.771159 | 0.0588792 |
| Chitin 1h | 308.03 | 35.9227 | 0.932171 | 0.120282 |
| Epi 1h | 289.686 | 26.9932 | 0.914112 | 0.0719879 |
| SA 1h | 258.934 | 15.3131 | 0.694495 | 0.0410548 |
| Me-JA 1h | 95.2147 | 14.2093 | 0.314865 | 0.050519 |
| Control 6h | 936.297 | 211.805 | 2.43057 | 0.417042 |
| ABA 6h | 116.518 | 7.6585 | 0.30142 | 0.0197875 |
| ACC 6h | 1305.15 | 193.346 | 3.07192 | 0.177621 |
| BABA 6h | 761.508 | 74.1164 | 1.85607 | 0.119784 |
| Chitin 6h | 683.597 | 39.6992 | 1.76821 | 0.102581 |
| Epi 6h | 810.08 | 73.9623 | 1.96358 | 0.291148 |
| SA 6h | 439.131 | 55.9827 | 1.20289 | 0.0702739 |
| Me-JA 6h | 369.707 | 50.8675 | 1.00325 | 0.0588069 |
Source Transcript PGSC0003DMT400055166 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G10140.1 | +1 | 3e-07 | 49 | 49/158 (31%) | Uncharacterised conserved protein UCP031279 | chr1:3322970-3323473 REVERSE LENGTH=167 |
