Probe CUST_44036_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_44036_PI426222305 | JHI_St_60k_v1 | DMT400016500 | CAAAGGATGGATCAGCAGAGCAATCTTCAGTTTCATGTCAAGTAATTTTGACAAATTTGT |
All Microarray Probes Designed to Gene DMG400006444
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_44036_PI426222305 | JHI_St_60k_v1 | DMT400016500 | CAAAGGATGGATCAGCAGAGCAATCTTCAGTTTCATGTCAAGTAATTTTGACAAATTTGT |
Microarray Signals from CUST_44036_PI426222305
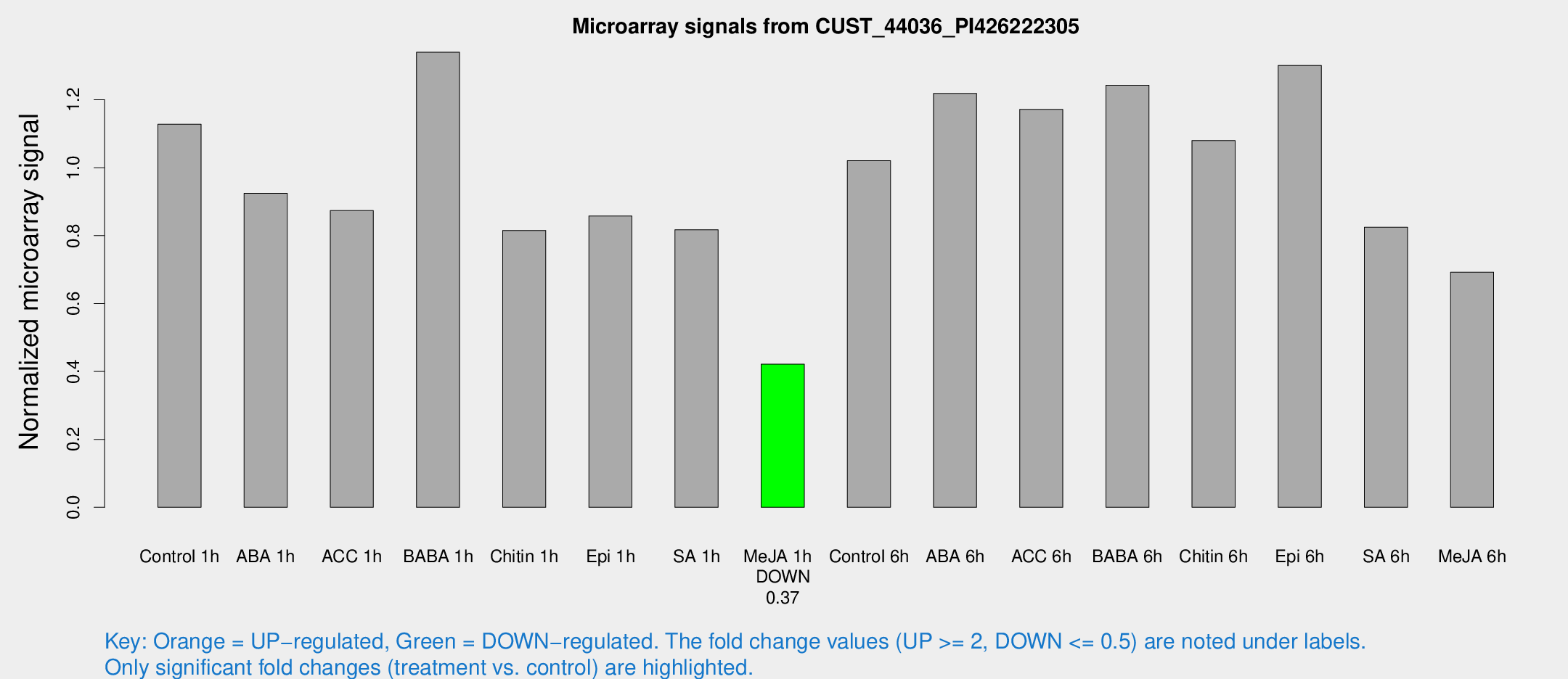
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 505.459 | 77.0992 | 1.1282 | 0.173261 |
| ABA 1h | 373.821 | 78.7574 | 0.924791 | 0.133208 |
| ACC 1h | 449.696 | 143.579 | 0.873635 | 0.305668 |
| BABA 1h | 566.395 | 33.6066 | 1.34013 | 0.143908 |
| Chitin 1h | 343.09 | 79.7892 | 0.815274 | 0.243963 |
| Epi 1h | 360.051 | 103.404 | 0.857789 | 0.308779 |
| SA 1h | 370.13 | 34.5178 | 0.817391 | 0.129803 |
| Me-JA 1h | 151.04 | 10.6578 | 0.421361 | 0.0258998 |
| Control 6h | 562.107 | 219.743 | 1.02053 | 0.52174 |
| ABA 6h | 593.409 | 134.75 | 1.21873 | 0.3309 |
| ACC 6h | 626.404 | 156.108 | 1.17172 | 0.164135 |
| BABA 6h | 628.644 | 112.525 | 1.24316 | 0.181818 |
| Chitin 6h | 502.472 | 29.2683 | 1.07986 | 0.062882 |
| Epi 6h | 659.278 | 113.971 | 1.30111 | 0.208624 |
| SA 6h | 367.92 | 65.1624 | 0.824554 | 0.0776054 |
| Me-JA 6h | 340.513 | 113.564 | 0.692314 | 0.231805 |
Source Transcript PGSC0003DMT400016500 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G14750.1 | +1 | 2e-65 | 218 | 157/325 (48%) | IQ-domain 19 | chr4:8470449-8471903 FORWARD LENGTH=387 |
