Probe CUST_4391_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_4391_PI426222305 | JHI_St_60k_v1 | DMT400020922 | GGCCAACATGGAGCGATCGTTACAGTTATGGATTTGATAGAATTCAGTAACTTTATATGT |
All Microarray Probes Designed to Gene DMG400008100
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_4391_PI426222305 | JHI_St_60k_v1 | DMT400020922 | GGCCAACATGGAGCGATCGTTACAGTTATGGATTTGATAGAATTCAGTAACTTTATATGT |
Microarray Signals from CUST_4391_PI426222305
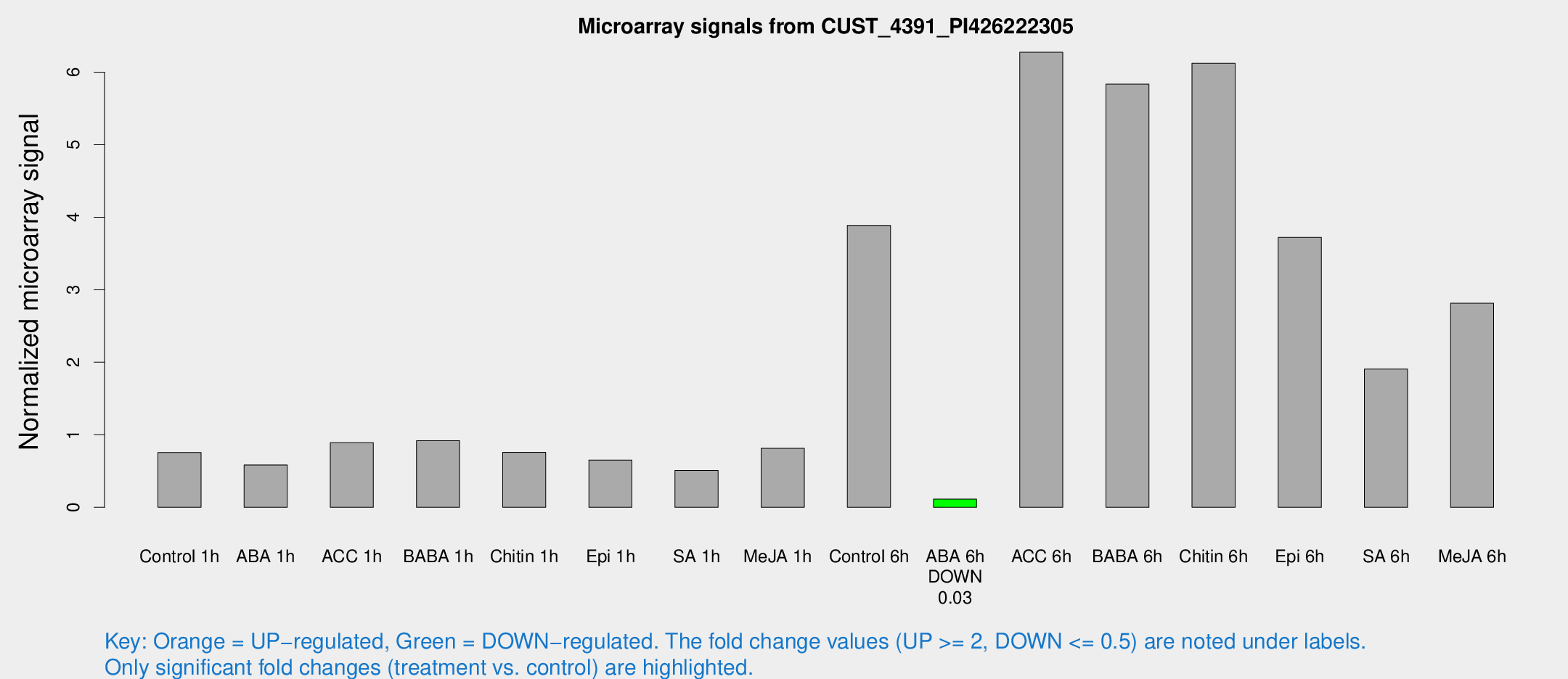
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 51.9793 | 9.45193 | 0.756732 | 0.11907 |
| ABA 1h | 36.0505 | 7.67296 | 0.584406 | 0.108562 |
| ACC 1h | 61.5556 | 8.49393 | 0.891576 | 0.0980377 |
| BABA 1h | 59.7631 | 8.50388 | 0.918797 | 0.0737208 |
| Chitin 1h | 45.456 | 4.23592 | 0.75877 | 0.115743 |
| Epi 1h | 39.2238 | 9.43506 | 0.650987 | 0.132733 |
| SA 1h | 34.6703 | 3.59253 | 0.508845 | 0.0529901 |
| Me-JA 1h | 46.2037 | 9.75918 | 0.812529 | 0.203378 |
| Control 6h | 288.293 | 82.5164 | 3.88767 | 1.09411 |
| ABA 6h | 8.82432 | 3.28281 | 0.1127 | 0.0528937 |
| ACC 6h | 542.21 | 206.327 | 6.27456 | 1.43361 |
| BABA 6h | 518.731 | 218.485 | 5.83377 | 2.5943 |
| Chitin 6h | 452.122 | 90.9701 | 6.12128 | 1.30859 |
| Epi 6h | 279.073 | 22.8926 | 3.7223 | 0.603107 |
| SA 6h | 124.722 | 7.93286 | 1.9057 | 0.315038 |
| Me-JA 6h | 212.3 | 68.2924 | 2.81317 | 0.948926 |
Source Transcript PGSC0003DMT400020922 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
