Probe CUST_43710_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43710_PI426222305 | JHI_St_60k_v1 | DMT400004871 | GGCAGTGAAATGTAGACTCAATTAACCCCTTGTTTTCCTTATATTATGTTCTATCCTTCA |
All Microarray Probes Designed to Gene DMG400001936
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43662_PI426222305 | JHI_St_60k_v1 | DMT400004870 | GCGACATCTTATACTACACTCCTTTTTCCCATCGTCTTCCAGTAGCTATTGGTTTTTACT |
| CUST_43710_PI426222305 | JHI_St_60k_v1 | DMT400004871 | GGCAGTGAAATGTAGACTCAATTAACCCCTTGTTTTCCTTATATTATGTTCTATCCTTCA |
Microarray Signals from CUST_43710_PI426222305
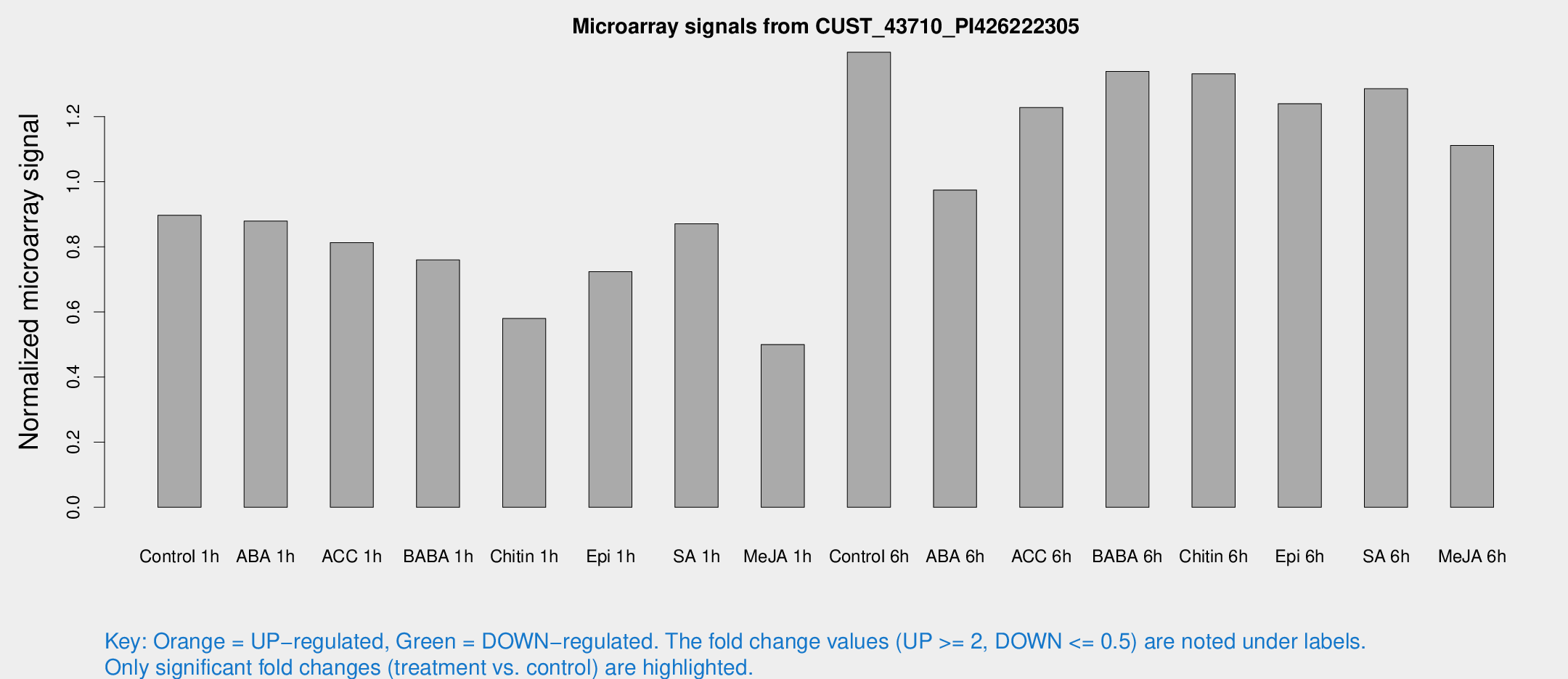
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 682.008 | 93.054 | 0.896973 | 0.0803679 |
| ABA 1h | 583.068 | 34.0244 | 0.879196 | 0.0509471 |
| ACC 1h | 689.883 | 187.719 | 0.812942 | 0.224451 |
| BABA 1h | 554.031 | 60.7002 | 0.759687 | 0.0440923 |
| Chitin 1h | 391.566 | 27.5362 | 0.579807 | 0.0337674 |
| Epi 1h | 473.336 | 48.8119 | 0.72363 | 0.0663756 |
| SA 1h | 670.768 | 42.2307 | 0.870616 | 0.0504176 |
| Me-JA 1h | 306.341 | 21.0964 | 0.499844 | 0.0292827 |
| Control 6h | 1114.32 | 248.824 | 1.39743 | 0.241862 |
| ABA 6h | 777.267 | 51.2764 | 0.974441 | 0.0564033 |
| ACC 6h | 1066.87 | 125.674 | 1.22772 | 0.0971972 |
| BABA 6h | 1124.18 | 71.3383 | 1.33856 | 0.0773905 |
| Chitin 6h | 1059.26 | 61.2703 | 1.33142 | 0.0769874 |
| Epi 6h | 1054.04 | 104.004 | 1.23951 | 0.121748 |
| SA 6h | 953.489 | 58.7107 | 1.28535 | 0.100197 |
| Me-JA 6h | 861.738 | 174.383 | 1.11143 | 0.134173 |
Source Transcript PGSC0003DMT400004871 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G03810.1 | +2 | 1e-34 | 136 | 120/396 (30%) | 18S pre-ribosomal assembly protein gar2-related | chr2:1162703-1164286 FORWARD LENGTH=439 |
