Probe CUST_43688_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43688_PI426222305 | JHI_St_60k_v1 | DMT400004881 | TCTACCTTACCACTTCTTGCTCAAGCAAAACCAAGATTAGCTAGGGCCATGTATCTTTAA |
All Microarray Probes Designed to Gene DMG401001941
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43688_PI426222305 | JHI_St_60k_v1 | DMT400004881 | TCTACCTTACCACTTCTTGCTCAAGCAAAACCAAGATTAGCTAGGGCCATGTATCTTTAA |
Microarray Signals from CUST_43688_PI426222305
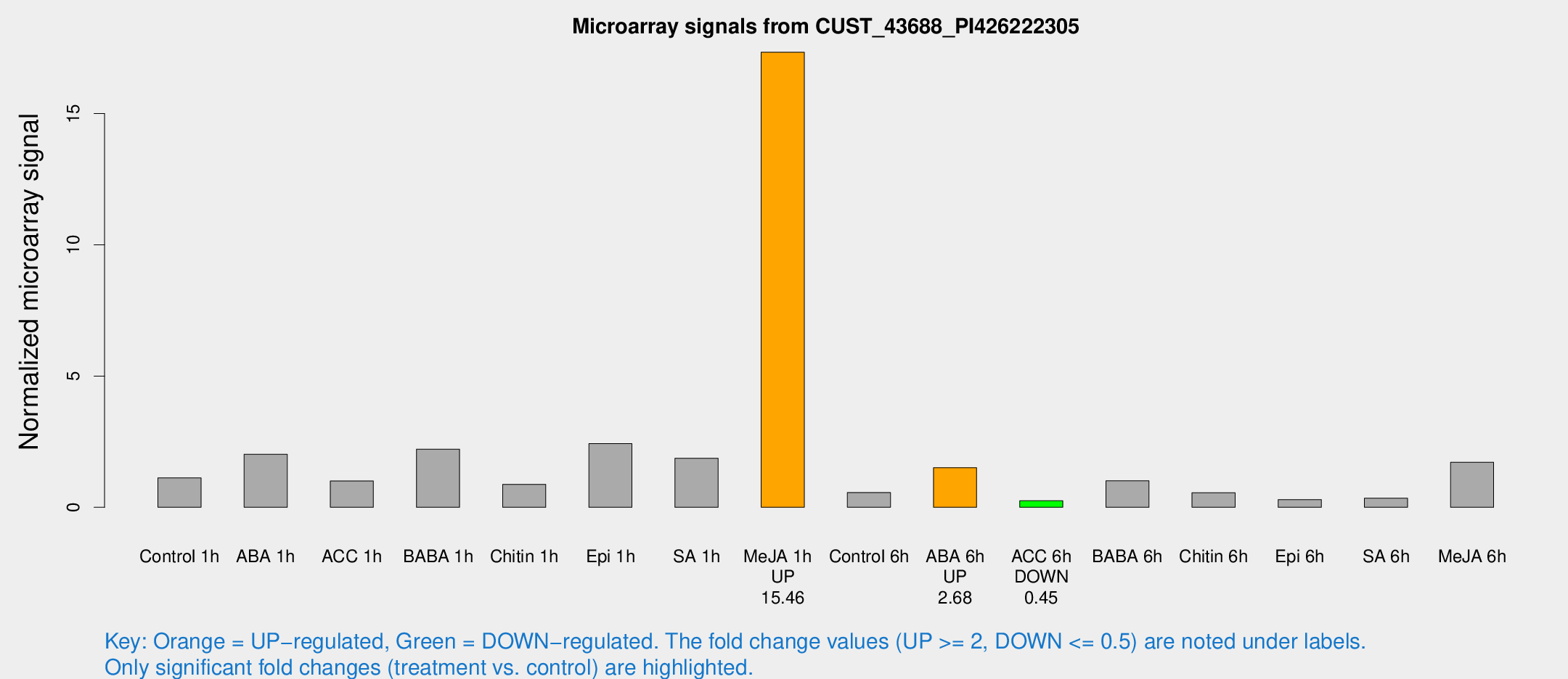
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 31.5508 | 7.7167 | 1.12118 | 0.33028 |
| ABA 1h | 67.368 | 39.3847 | 2.02273 | 1.36834 |
| ACC 1h | 29.1809 | 7.97173 | 1.00267 | 0.236168 |
| BABA 1h | 72.765 | 31.5841 | 2.21527 | 1.20622 |
| Chitin 1h | 21.9018 | 4.86167 | 0.875837 | 0.31598 |
| Epi 1h | 67.8325 | 25.8334 | 2.42563 | 1.41228 |
| SA 1h | 54.5461 | 16.4787 | 1.86865 | 0.67631 |
| Me-JA 1h | 371.668 | 24.4246 | 17.3363 | 1.54366 |
| Control 6h | 14.8745 | 3.99139 | 0.562661 | 0.152976 |
| ABA 6h | 42.0451 | 4.38282 | 1.51057 | 0.158813 |
| ACC 6h | 7.69384 | 4.55285 | 0.25179 | 0.146076 |
| BABA 6h | 29.6982 | 4.58091 | 1.00971 | 0.155385 |
| Chitin 6h | 16.2405 | 4.39687 | 0.556728 | 0.170634 |
| Epi 6h | 9.06139 | 4.82384 | 0.29192 | 0.157759 |
| SA 6h | 9.34509 | 4.17627 | 0.346063 | 0.170184 |
| Me-JA 6h | 53.906 | 22.542 | 1.71754 | 0.770295 |
Source Transcript PGSC0003DMT400004881 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G05260.1 | +1 | 1e-132 | 396 | 238/460 (52%) | cytochrome p450 79a2 | chr5:1559778-1561765 REVERSE LENGTH=523 |
