Probe CUST_43662_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43662_PI426222305 | JHI_St_60k_v1 | DMT400004870 | GCGACATCTTATACTACACTCCTTTTTCCCATCGTCTTCCAGTAGCTATTGGTTTTTACT |
All Microarray Probes Designed to Gene DMG400001936
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43662_PI426222305 | JHI_St_60k_v1 | DMT400004870 | GCGACATCTTATACTACACTCCTTTTTCCCATCGTCTTCCAGTAGCTATTGGTTTTTACT |
| CUST_43710_PI426222305 | JHI_St_60k_v1 | DMT400004871 | GGCAGTGAAATGTAGACTCAATTAACCCCTTGTTTTCCTTATATTATGTTCTATCCTTCA |
Microarray Signals from CUST_43662_PI426222305
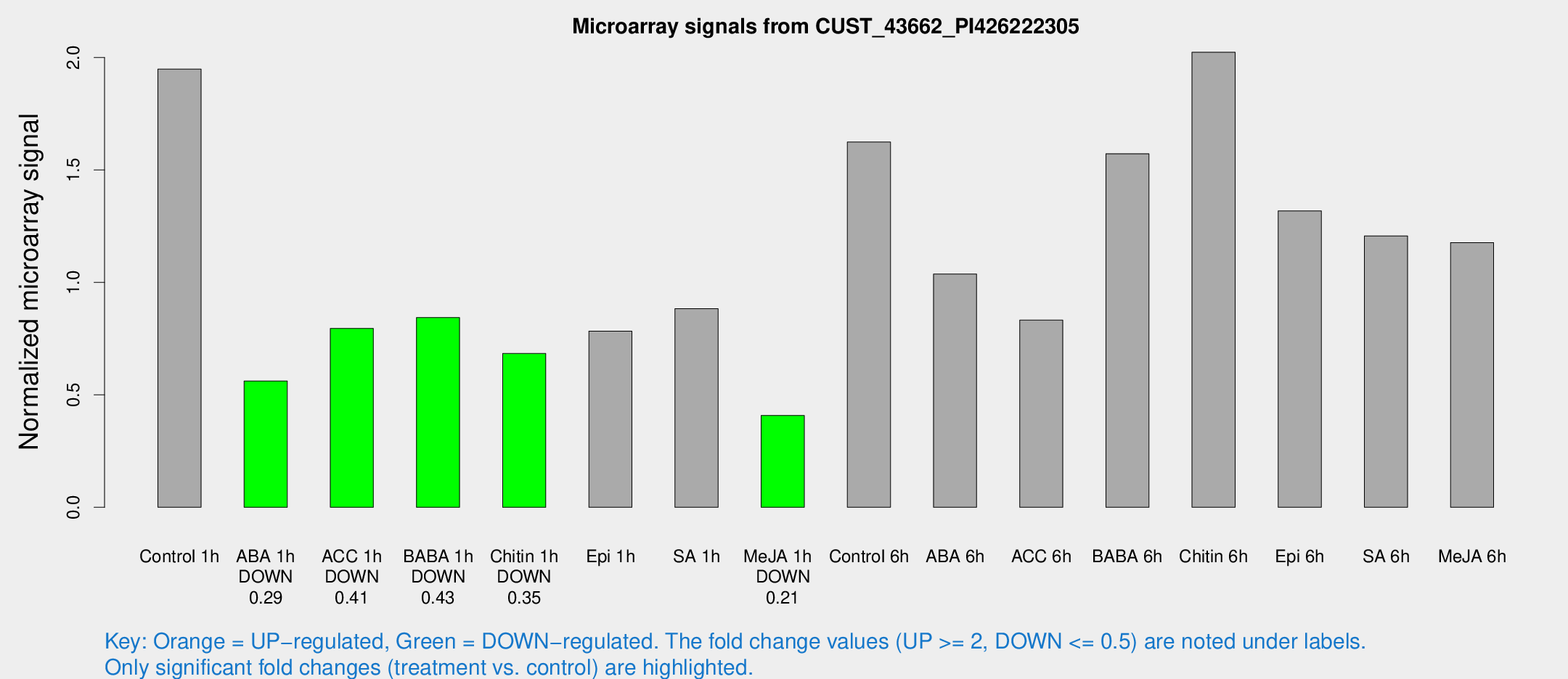
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 41.1354 | 4.35796 | 1.94853 | 0.207291 |
| ABA 1h | 11.7936 | 4.30835 | 0.561772 | 0.212801 |
| ACC 1h | 18.2548 | 4.71037 | 0.795085 | 0.267694 |
| BABA 1h | 17.4145 | 3.96544 | 0.843622 | 0.198919 |
| Chitin 1h | 13.1807 | 3.74108 | 0.683781 | 0.203067 |
| Epi 1h | 15.2526 | 4.16713 | 0.78319 | 0.222943 |
| SA 1h | 21.5281 | 7.48787 | 0.882884 | 0.413278 |
| Me-JA 1h | 7.06736 | 3.7763 | 0.407976 | 0.219705 |
| Control 6h | 34.2809 | 4.27846 | 1.62397 | 0.203998 |
| ABA 6h | 24.2338 | 4.86873 | 1.03689 | 0.25798 |
| ACC 6h | 21.6537 | 6.14132 | 0.832342 | 0.316126 |
| BABA 6h | 39.6285 | 10.8358 | 1.57197 | 0.399268 |
| Chitin 6h | 52.2473 | 18.944 | 2.0235 | 0.93109 |
| Epi 6h | 32.0451 | 5.04315 | 1.31812 | 0.277993 |
| SA 6h | 29.1175 | 10.8244 | 1.20668 | 0.560365 |
| Me-JA 6h | 25.279 | 4.16518 | 1.17716 | 0.196016 |
Source Transcript PGSC0003DMT400004870 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G03810.1 | +1 | 5e-23 | 71 | 83/356 (23%) | 18S pre-ribosomal assembly protein gar2-related | chr2:1162703-1164286 FORWARD LENGTH=439 |
