Probe CUST_43377_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43377_PI426222305 | JHI_St_60k_v1 | DMT400033027 | TTTGTGGTACTTTGTATGTATGCCCTATTTTCTGAATTGCATCCCTATGTGATAAGTTCC |
All Microarray Probes Designed to Gene DMG401012679
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43377_PI426222305 | JHI_St_60k_v1 | DMT400033027 | TTTGTGGTACTTTGTATGTATGCCCTATTTTCTGAATTGCATCCCTATGTGATAAGTTCC |
Microarray Signals from CUST_43377_PI426222305
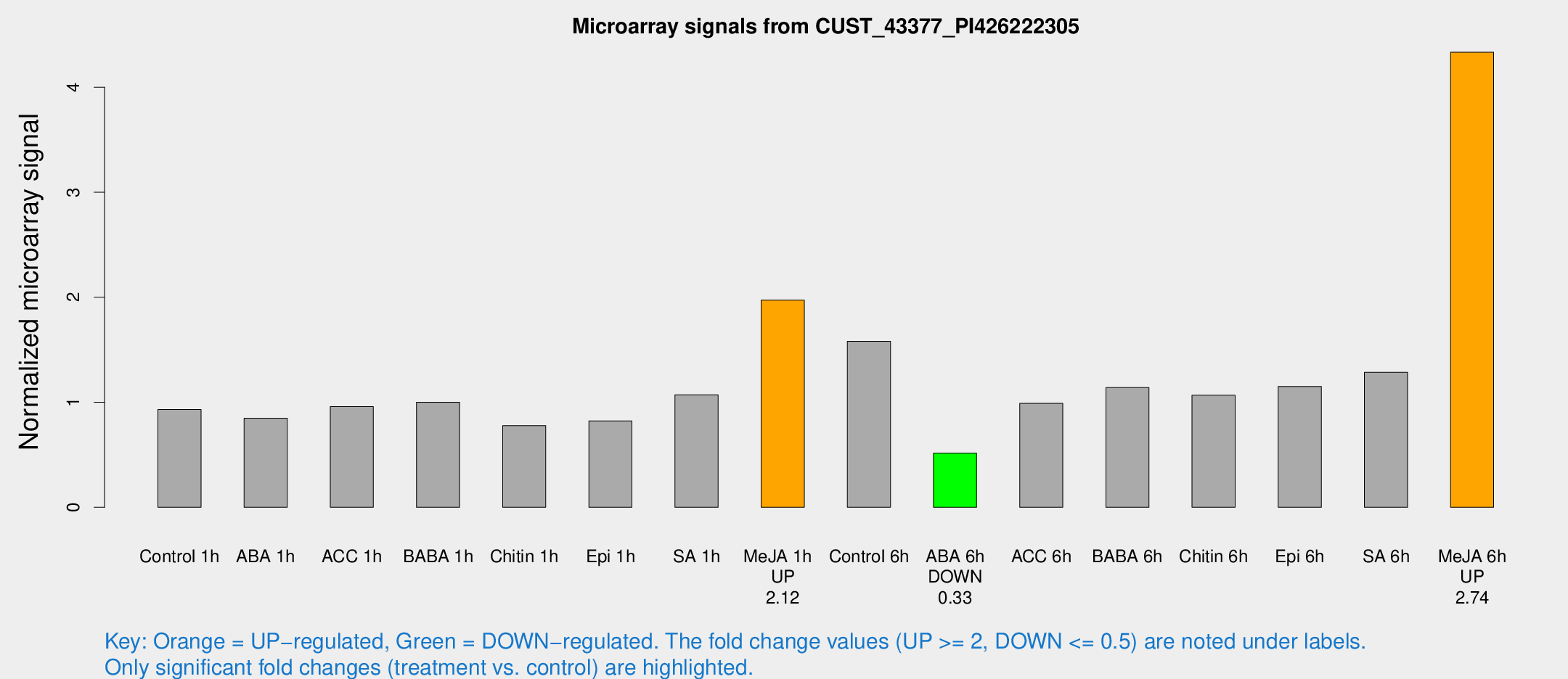
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1371.73 | 79.3311 | 0.930322 | 0.0537494 |
| ABA 1h | 1109.73 | 78.9472 | 0.847489 | 0.0489786 |
| ACC 1h | 1500.02 | 256.947 | 0.958678 | 0.106322 |
| BABA 1h | 1425.04 | 82.5817 | 1.00071 | 0.0783448 |
| Chitin 1h | 1044.33 | 121.567 | 0.778024 | 0.0869311 |
| Epi 1h | 1062.13 | 118.611 | 0.822393 | 0.0911665 |
| SA 1h | 1780.03 | 569.24 | 1.0713 | 0.360053 |
| Me-JA 1h | 2382.3 | 156.5 | 1.97216 | 0.117751 |
| Control 6h | 2474.39 | 614.031 | 1.58041 | 0.267631 |
| ABA 6h | 825.96 | 117.031 | 0.515879 | 0.089819 |
| ACC 6h | 1699.68 | 212.336 | 0.989249 | 0.185914 |
| BABA 6h | 1949.33 | 383.569 | 1.13952 | 0.272388 |
| Chitin 6h | 1741.02 | 362.521 | 1.06712 | 0.214962 |
| Epi 6h | 1935.24 | 213.345 | 1.15043 | 0.199534 |
| SA 6h | 1968.19 | 401.84 | 1.28523 | 0.150815 |
| Me-JA 6h | 6370.78 | 368.769 | 4.3324 | 0.576524 |
Source Transcript PGSC0003DMT400033027 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G25780.1 | +2 | 3e-70 | 223 | 125/202 (62%) | allene oxide cyclase 3 | chr3:9409362-9410409 FORWARD LENGTH=258 |
