Probe CUST_43280_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43280_PI426222305 | JHI_St_60k_v1 | DMT400064432 | CGATGTCCGGCATCTGTATTGGCATTCTCATGAATTAATTAGTTGAAATATATATCTGAG |
All Microarray Probes Designed to Gene DMG400025033
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43280_PI426222305 | JHI_St_60k_v1 | DMT400064432 | CGATGTCCGGCATCTGTATTGGCATTCTCATGAATTAATTAGTTGAAATATATATCTGAG |
Microarray Signals from CUST_43280_PI426222305
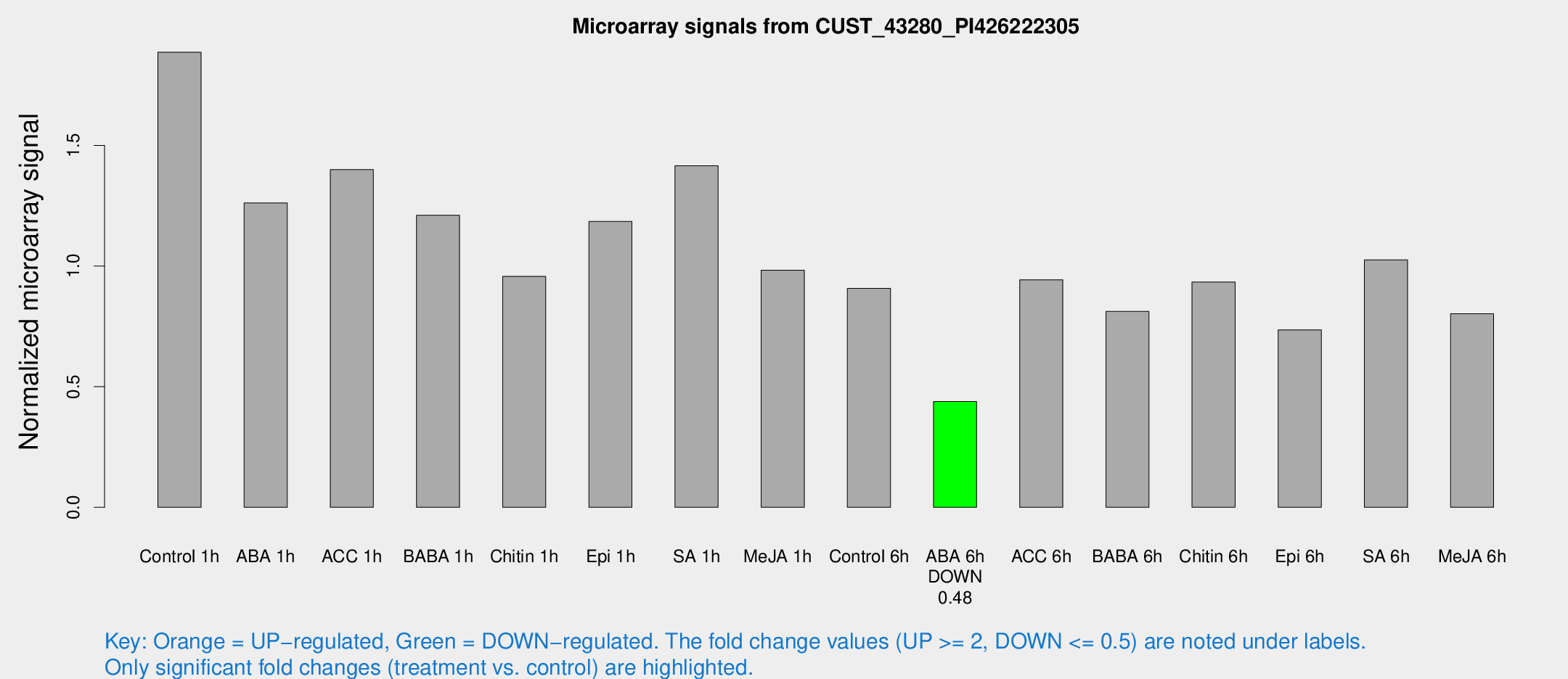
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 228.411 | 27.0749 | 1.88652 | 0.113478 |
| ABA 1h | 133.736 | 8.52511 | 1.2615 | 0.141706 |
| ACC 1h | 179.016 | 33.8569 | 1.39991 | 0.195013 |
| BABA 1h | 142.899 | 21.2391 | 1.21082 | 0.0783024 |
| Chitin 1h | 109.178 | 25.6767 | 0.957229 | 0.211807 |
| Epi 1h | 123.079 | 9.04207 | 1.18488 | 0.0770585 |
| SA 1h | 174.873 | 14.5585 | 1.41626 | 0.163691 |
| Me-JA 1h | 96.9446 | 10.7325 | 0.982524 | 0.104184 |
| Control 6h | 109.213 | 8.1188 | 0.907667 | 0.0618491 |
| ABA 6h | 55.6956 | 5.10186 | 0.438483 | 0.0402986 |
| ACC 6h | 130.56 | 13.7625 | 0.943231 | 0.0637957 |
| BABA 6h | 109.325 | 9.18467 | 0.812081 | 0.0902548 |
| Chitin 6h | 118.881 | 8.06287 | 0.933616 | 0.0643737 |
| Epi 6h | 100.504 | 11.7716 | 0.735836 | 0.061061 |
| SA 6h | 122.166 | 11.4301 | 1.02588 | 0.106192 |
| Me-JA 6h | 96.7853 | 12.0617 | 0.802282 | 0.0602074 |
Source Transcript PGSC0003DMT400064432 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G27470.1 | +3 | 2e-40 | 147 | 77/195 (39%) | RING membrane-anchor 3 | chr4:13735576-13736307 FORWARD LENGTH=243 |
