Probe CUST_43211_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43211_PI426222305 | JHI_St_60k_v1 | DMT400042701 | TTTAGACGAGGCGGAGTACACAATTGTAAAGAACATCATACAGATTGAATACTACTCTAT |
All Microarray Probes Designed to Gene DMG400016570
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43211_PI426222305 | JHI_St_60k_v1 | DMT400042701 | TTTAGACGAGGCGGAGTACACAATTGTAAAGAACATCATACAGATTGAATACTACTCTAT |
Microarray Signals from CUST_43211_PI426222305
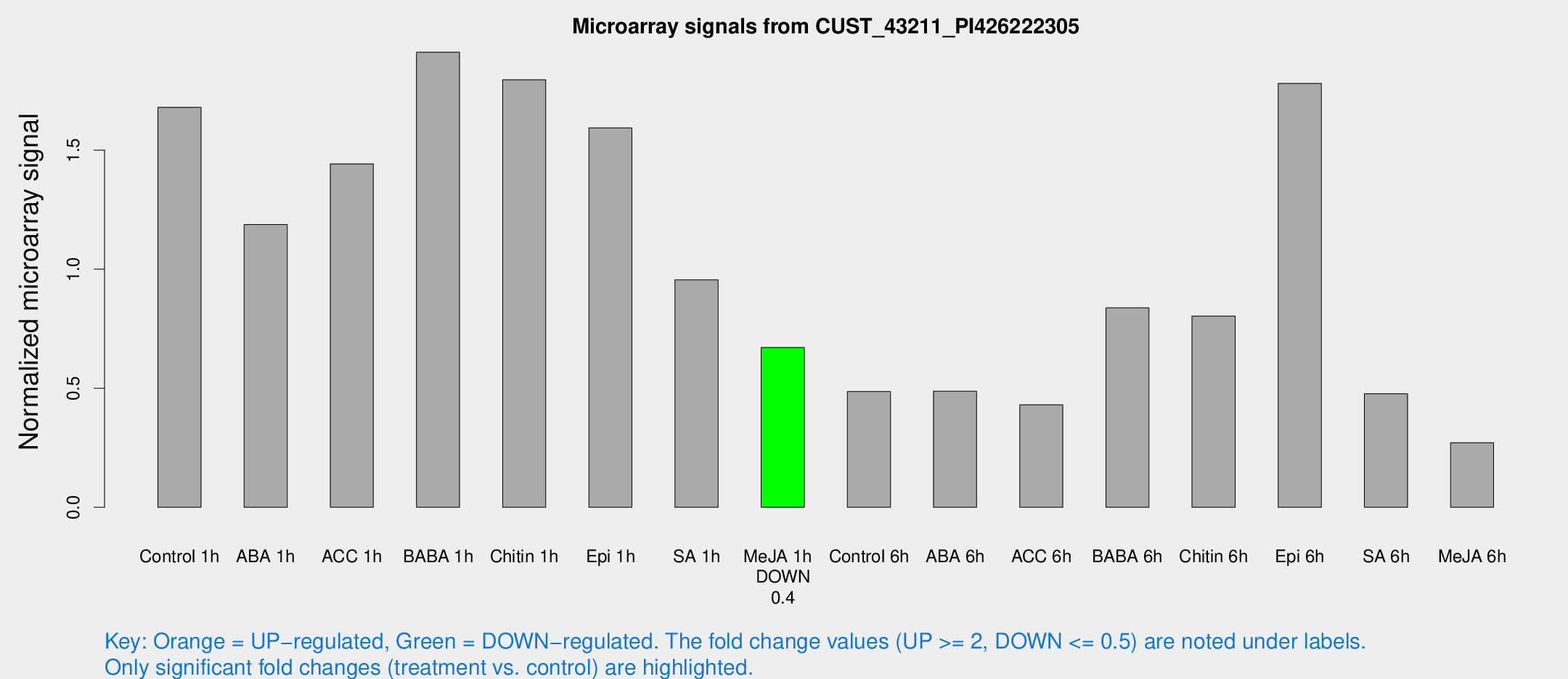
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 244.987 | 18.1107 | 1.67943 | 0.0999688 |
| ABA 1h | 166.492 | 47.5182 | 1.18745 | 0.291226 |
| ACC 1h | 226.75 | 47.596 | 1.442 | 0.244381 |
| BABA 1h | 298.481 | 96.9073 | 1.9109 | 0.505729 |
| Chitin 1h | 242.702 | 46.7109 | 1.79552 | 0.426036 |
| Epi 1h | 203.164 | 26.0524 | 1.59345 | 0.154786 |
| SA 1h | 142.691 | 8.97454 | 0.955161 | 0.108471 |
| Me-JA 1h | 79.8111 | 5.87432 | 0.671031 | 0.0493015 |
| Control 6h | 87.1608 | 31.5996 | 0.486396 | 0.228907 |
| ABA 6h | 84.263 | 29.6657 | 0.487841 | 0.206329 |
| ACC 6h | 85.4459 | 37.5563 | 0.430208 | 0.122147 |
| BABA 6h | 151.097 | 51.3496 | 0.837739 | 0.258718 |
| Chitin 6h | 134.462 | 39.1463 | 0.80315 | 0.232176 |
| Epi 6h | 297.609 | 45.3652 | 1.77977 | 0.325066 |
| SA 6h | 72.5851 | 15.6956 | 0.47726 | 0.173261 |
| Me-JA 6h | 46.7616 | 18.9546 | 0.271623 | 0.116574 |
Source Transcript PGSC0003DMT400042701 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G29500.1 | +2 | 8e-33 | 117 | 67/122 (55%) | SAUR-like auxin-responsive protein family | chr1:10321290-10321697 FORWARD LENGTH=135 |
