Probe CUST_43206_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43206_PI426222305 | JHI_St_60k_v1 | DMT400042691 | GCAGCTTCTTGTTTGTTGAGTTAAGGTCCACAATTTTTTGTACATGGTAGCTCAAAAATA |
All Microarray Probes Designed to Gene DMG400016562
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43206_PI426222305 | JHI_St_60k_v1 | DMT400042691 | GCAGCTTCTTGTTTGTTGAGTTAAGGTCCACAATTTTTTGTACATGGTAGCTCAAAAATA |
Microarray Signals from CUST_43206_PI426222305
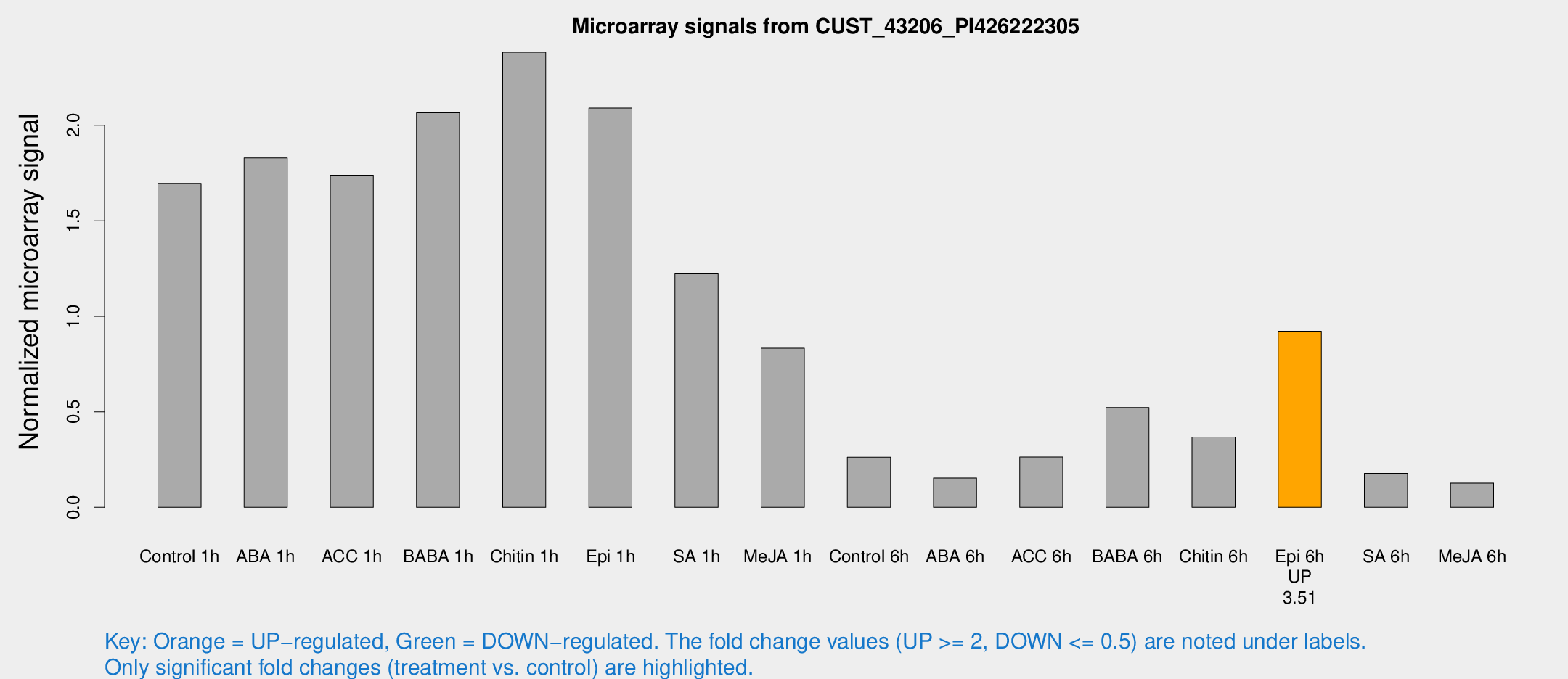
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 217.412 | 23.3492 | 1.69586 | 0.100776 |
| ABA 1h | 218.313 | 50.4603 | 1.82907 | 0.371385 |
| ACC 1h | 226.272 | 13.5255 | 1.73846 | 0.150066 |
| BABA 1h | 264.327 | 54.0465 | 2.0653 | 0.260858 |
| Chitin 1h | 280.648 | 53.3342 | 2.38245 | 0.525138 |
| Epi 1h | 235.632 | 38.4341 | 2.08986 | 0.31363 |
| SA 1h | 159.206 | 9.68276 | 1.22188 | 0.0868194 |
| Me-JA 1h | 91.8691 | 22.6167 | 0.832855 | 0.14626 |
| Control 6h | 38.8113 | 12.5867 | 0.262621 | 0.0935125 |
| ABA 6h | 25.1859 | 10.6846 | 0.153529 | 0.0891871 |
| ACC 6h | 45.8823 | 20.3016 | 0.262899 | 0.0817819 |
| BABA 6h | 88.0332 | 38.2193 | 0.522142 | 0.219928 |
| Chitin 6h | 57.247 | 22.7689 | 0.367266 | 0.149392 |
| Epi 6h | 134.313 | 19.408 | 0.922414 | 0.134229 |
| SA 6h | 22.5404 | 3.50695 | 0.177883 | 0.0285502 |
| Me-JA 6h | 23.4872 | 13.1627 | 0.126704 | 0.103539 |
Source Transcript PGSC0003DMT400042691 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G29510.1 | +1 | 5e-40 | 136 | 79/147 (54%) | SAUR-like auxin-responsive protein family | chr1:10322683-10323114 FORWARD LENGTH=143 |
