Probe CUST_42887_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42887_PI426222305 | JHI_St_60k_v1 | DMT400058486 | GGTGTTCATCTTGTTAAGAATATGCCTAATGAAGCACTTAATTGTGGTTTGACTAGGAAA |
All Microarray Probes Designed to Gene DMG400022713
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42911_PI426222305 | JHI_St_60k_v1 | DMT400058487 | GGTGTTCATCTTGTTAAGAATATGCCTAATGAAGCACTTAATTGTGGTTTGACTAGGAAA |
| CUST_42887_PI426222305 | JHI_St_60k_v1 | DMT400058486 | GGTGTTCATCTTGTTAAGAATATGCCTAATGAAGCACTTAATTGTGGTTTGACTAGGAAA |
Microarray Signals from CUST_42887_PI426222305
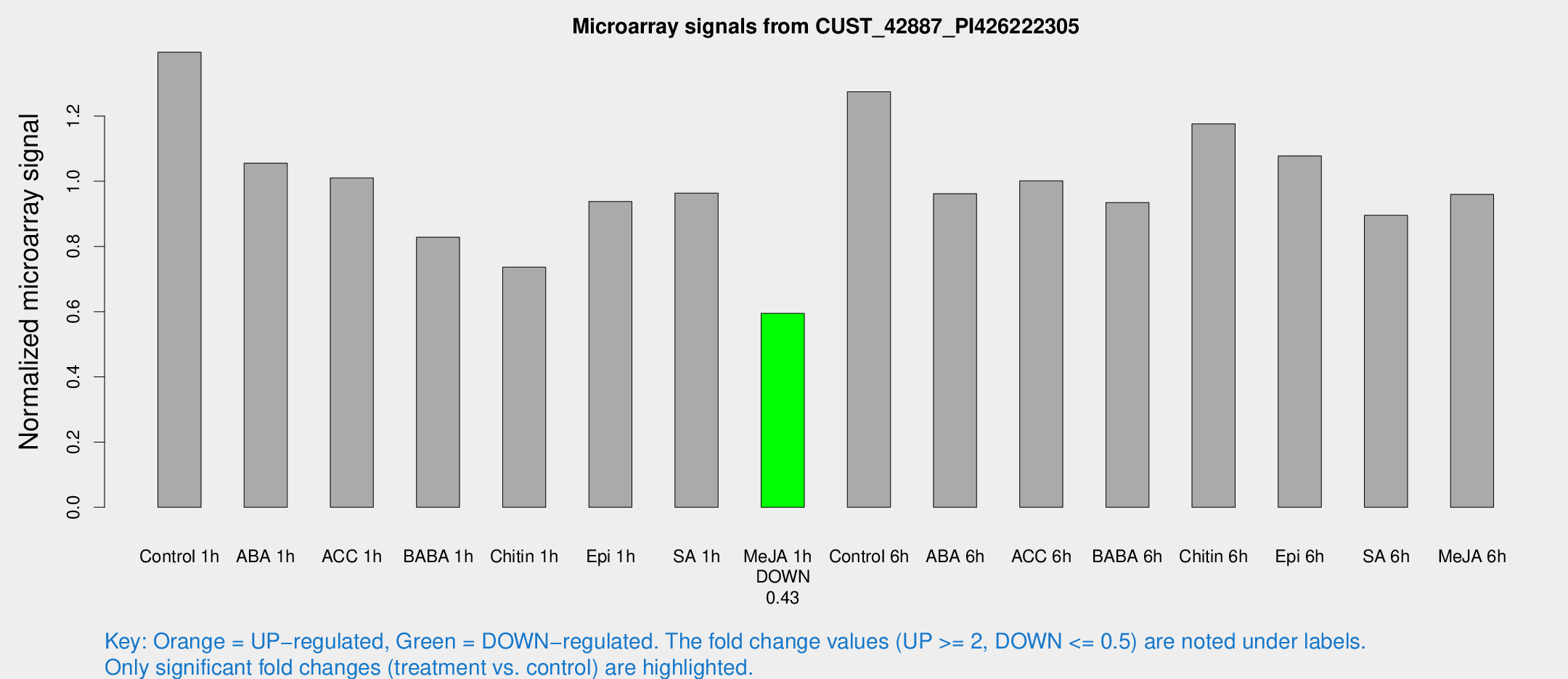
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 741.196 | 141.106 | 1.39569 | 0.193446 |
| ABA 1h | 480.233 | 27.9182 | 1.0554 | 0.0612402 |
| ACC 1h | 573.849 | 139.061 | 1.01025 | 0.221081 |
| BABA 1h | 440.777 | 104.788 | 0.828346 | 0.151523 |
| Chitin 1h | 340.559 | 19.9052 | 0.736591 | 0.0429884 |
| Epi 1h | 422.509 | 50.786 | 0.937916 | 0.0840026 |
| SA 1h | 515.209 | 60.0795 | 0.963699 | 0.0559258 |
| Me-JA 1h | 252.409 | 27.6038 | 0.594553 | 0.0350534 |
| Control 6h | 693.891 | 151.26 | 1.2743 | 0.208955 |
| ABA 6h | 531.933 | 61.3978 | 0.961843 | 0.0562693 |
| ACC 6h | 607.831 | 98.1317 | 1.00142 | 0.134501 |
| BABA 6h | 555.67 | 105.424 | 0.934733 | 0.178937 |
| Chitin 6h | 647.694 | 57.8854 | 1.17581 | 0.0681604 |
| Epi 6h | 623.974 | 36.1954 | 1.07771 | 0.0878964 |
| SA 6h | 514.562 | 151.828 | 0.895567 | 0.245839 |
| Me-JA 6h | 518.27 | 110.161 | 0.959599 | 0.160631 |
Source Transcript PGSC0003DMT400058486 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G21105.1 | +1 | 1e-173 | 504 | 240/383 (63%) | Plant L-ascorbate oxidase | chr5:7172727-7177409 FORWARD LENGTH=588 |
