Probe CUST_42676_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42676_PI426222305 | JHI_St_60k_v1 | DMT400052409 | GAAATGTGCCCCACCGAAATGAAACTTGAAATCATGAAGGGCAAAAAGGTTTTTTAAAAA |
All Microarray Probes Designed to Gene DMG400020343
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42676_PI426222305 | JHI_St_60k_v1 | DMT400052409 | GAAATGTGCCCCACCGAAATGAAACTTGAAATCATGAAGGGCAAAAAGGTTTTTTAAAAA |
Microarray Signals from CUST_42676_PI426222305
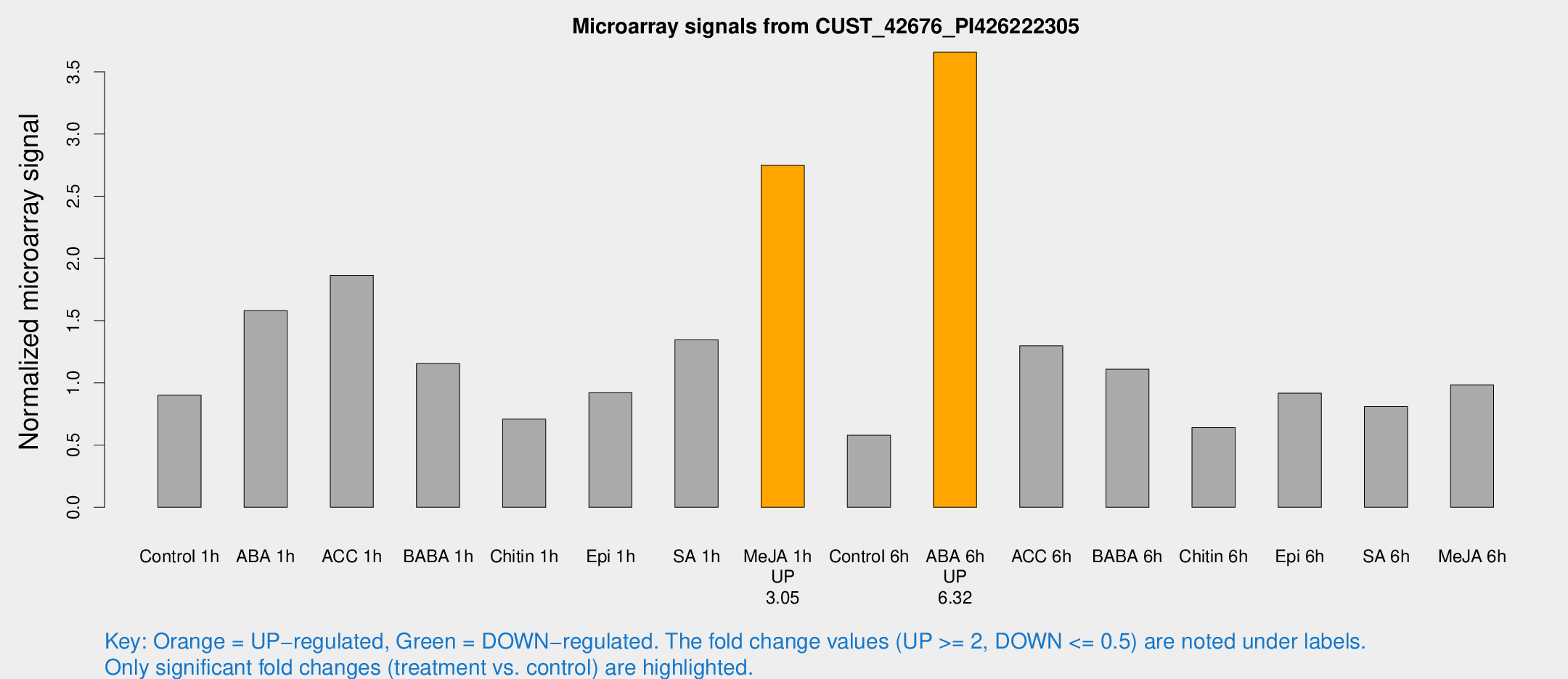
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1801 | 248.815 | 0.900855 | 0.0633048 |
| ABA 1h | 3101.14 | 908.91 | 1.58096 | 0.545085 |
| ACC 1h | 4335.2 | 1405.07 | 1.86509 | 0.67131 |
| BABA 1h | 2281.54 | 454.44 | 1.15553 | 0.138157 |
| Chitin 1h | 1277.97 | 174.888 | 0.709098 | 0.0724234 |
| Epi 1h | 1576.06 | 141.622 | 0.920422 | 0.092317 |
| SA 1h | 2860 | 658.332 | 1.34523 | 0.288971 |
| Me-JA 1h | 4538.45 | 764.624 | 2.74842 | 0.245109 |
| Control 6h | 1279.38 | 408.455 | 0.578726 | 0.158888 |
| ABA 6h | 7723.52 | 801.235 | 3.65781 | 0.228203 |
| ACC 6h | 3035.98 | 606.378 | 1.2976 | 0.123904 |
| BABA 6h | 2666.36 | 827.071 | 1.10993 | 0.316269 |
| Chitin 6h | 1350.06 | 119.328 | 0.641094 | 0.0669546 |
| Epi 6h | 2093.77 | 383.766 | 0.916979 | 0.16175 |
| SA 6h | 1612.25 | 239.987 | 0.809566 | 0.0467932 |
| Me-JA 6h | 1954.92 | 263.546 | 0.982404 | 0.117624 |
Source Transcript PGSC0003DMT400052409 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G01210.1 | +3 | 0.0 | 598 | 309/473 (65%) | HXXXD-type acyl-transferase family protein | chr5:84554-85981 FORWARD LENGTH=475 |
