Probe CUST_42667_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42667_PI426222305 | JHI_St_60k_v1 | DMT400052413 | CACCTCCAAAAGATCTTCCAAAATGCTAAAAAGGCTCTTTTGAAAGGAGTTGAATACAAA |
All Microarray Probes Designed to Gene DMG400020345
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42667_PI426222305 | JHI_St_60k_v1 | DMT400052413 | CACCTCCAAAAGATCTTCCAAAATGCTAAAAAGGCTCTTTTGAAAGGAGTTGAATACAAA |
Microarray Signals from CUST_42667_PI426222305
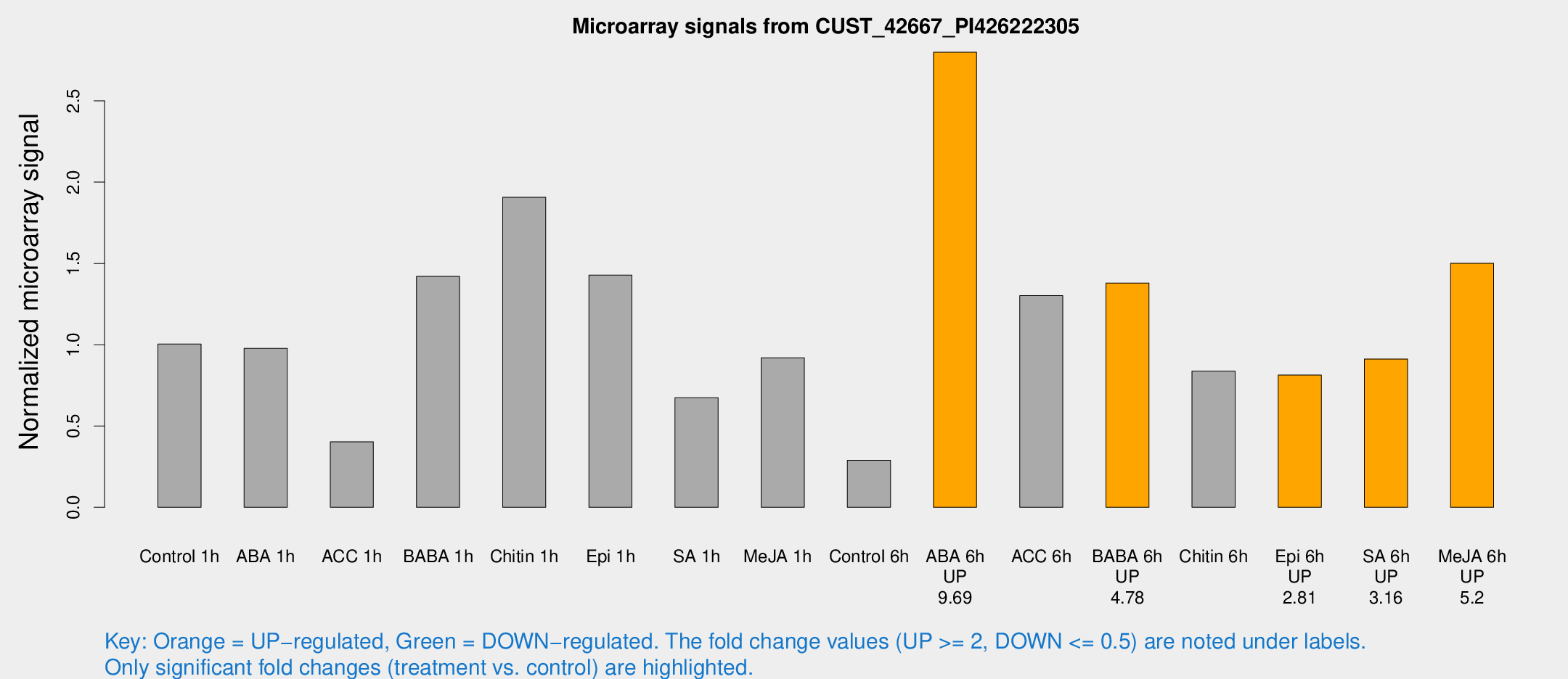
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 27.3935 | 4.90513 | 1.00384 | 0.162081 |
| ABA 1h | 24.258 | 5.69048 | 0.977347 | 0.18862 |
| ACC 1h | 13.4116 | 6.35271 | 0.402887 | 0.191612 |
| BABA 1h | 38.4219 | 9.38436 | 1.42039 | 0.540176 |
| Chitin 1h | 45.4092 | 4.6044 | 1.90706 | 0.192834 |
| Epi 1h | 38.0684 | 15.6783 | 1.42761 | 0.56054 |
| SA 1h | 23.4752 | 9.69585 | 0.674059 | 0.420042 |
| Me-JA 1h | 23.8754 | 8.20195 | 0.919675 | 0.653226 |
| Control 6h | 7.71898 | 3.96091 | 0.2889 | 0.150228 |
| ABA 6h | 79.3998 | 8.54844 | 2.79894 | 0.236758 |
| ACC 6h | 56.1414 | 23.228 | 1.30185 | 1.12327 |
| BABA 6h | 45.1276 | 13.4431 | 1.3795 | 0.541285 |
| Chitin 6h | 31.918 | 15.464 | 0.837628 | 0.606281 |
| Epi 6h | 25.5208 | 5.47188 | 0.813021 | 0.161819 |
| SA 6h | 24.0254 | 4.37481 | 0.912024 | 0.169267 |
| Me-JA 6h | 41.875 | 9.71914 | 1.50089 | 0.549157 |
Source Transcript PGSC0003DMT400052413 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G38080.1 | +2 | 0.0 | 858 | 406/543 (75%) | Laccase/Diphenol oxidase family protein | chr2:15934540-15937352 FORWARD LENGTH=558 |
