Probe CUST_42605_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42605_PI426222305 | JHI_St_60k_v1 | DMT400026782 | CCACTTCTACAACAATATCAGTGTGATATCGATCGGATTATTTCGCTTCATGTAAGTTTT |
All Microarray Probes Designed to Gene DMG400010342
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42560_PI426222305 | JHI_St_60k_v1 | DMT400026783 | GTGTGATATCGATCGGATTATTTCGCTTCATACTAAGAAAGTAAGGATGGAATTGGAAGA |
| CUST_42605_PI426222305 | JHI_St_60k_v1 | DMT400026782 | CCACTTCTACAACAATATCAGTGTGATATCGATCGGATTATTTCGCTTCATGTAAGTTTT |
Microarray Signals from CUST_42605_PI426222305
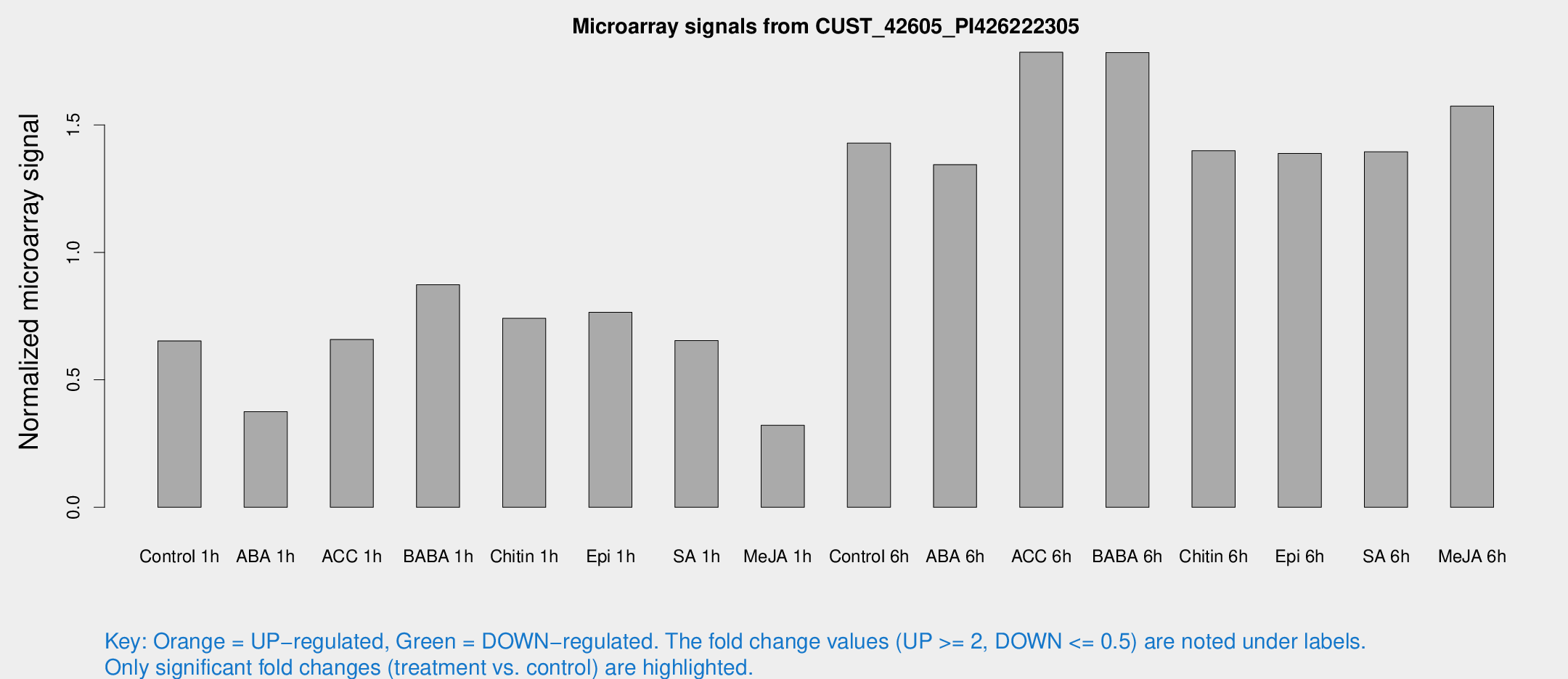
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 341.948 | 47.1306 | 0.652659 | 0.0766647 |
| ABA 1h | 174.476 | 25.7088 | 0.375096 | 0.031566 |
| ACC 1h | 420.417 | 145.426 | 0.658295 | 0.304182 |
| BABA 1h | 446.202 | 77.2206 | 0.87309 | 0.0829312 |
| Chitin 1h | 345.69 | 32.6886 | 0.741485 | 0.0447109 |
| Epi 1h | 352.905 | 62.103 | 0.764732 | 0.143949 |
| SA 1h | 351.305 | 43.4664 | 0.654056 | 0.0567022 |
| Me-JA 1h | 135.208 | 8.50768 | 0.321809 | 0.0349333 |
| Control 6h | 805.036 | 218.183 | 1.42865 | 0.340339 |
| ABA 6h | 751.559 | 107.931 | 1.34413 | 0.142146 |
| ACC 6h | 1067.41 | 125.527 | 1.78506 | 0.103283 |
| BABA 6h | 1067.95 | 201.18 | 1.78385 | 0.29798 |
| Chitin 6h | 770.313 | 60.3388 | 1.39859 | 0.116665 |
| Epi 6h | 829.911 | 152.879 | 1.38784 | 0.25311 |
| SA 6h | 731.446 | 126.008 | 1.39451 | 0.425729 |
| Me-JA 6h | 851.372 | 180.382 | 1.57395 | 0.28278 |
Source Transcript PGSC0003DMT400026782 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G79110.1 | +2 | 1e-63 | 212 | 114/201 (57%) | zinc ion binding | chr1:29759345-29760586 FORWARD LENGTH=358 |
