Probe CUST_42503_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42503_PI426222305 | JHI_St_60k_v1 | DMT400079243 | CTCCACAAAAGGACGTCTACATGGTGATATAATTAAGAGGACTGGCTGGGTTATAGGAAT |
All Microarray Probes Designed to Gene DMG400030843
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42503_PI426222305 | JHI_St_60k_v1 | DMT400079243 | CTCCACAAAAGGACGTCTACATGGTGATATAATTAAGAGGACTGGCTGGGTTATAGGAAT |
| CUST_42494_PI426222305 | JHI_St_60k_v1 | DMT400079244 | TGATATAATTAAGAGGAAGTTTGTGGTATGCTGCAATGGTTCCTTGCCTACTCCTCCCTC |
Microarray Signals from CUST_42503_PI426222305
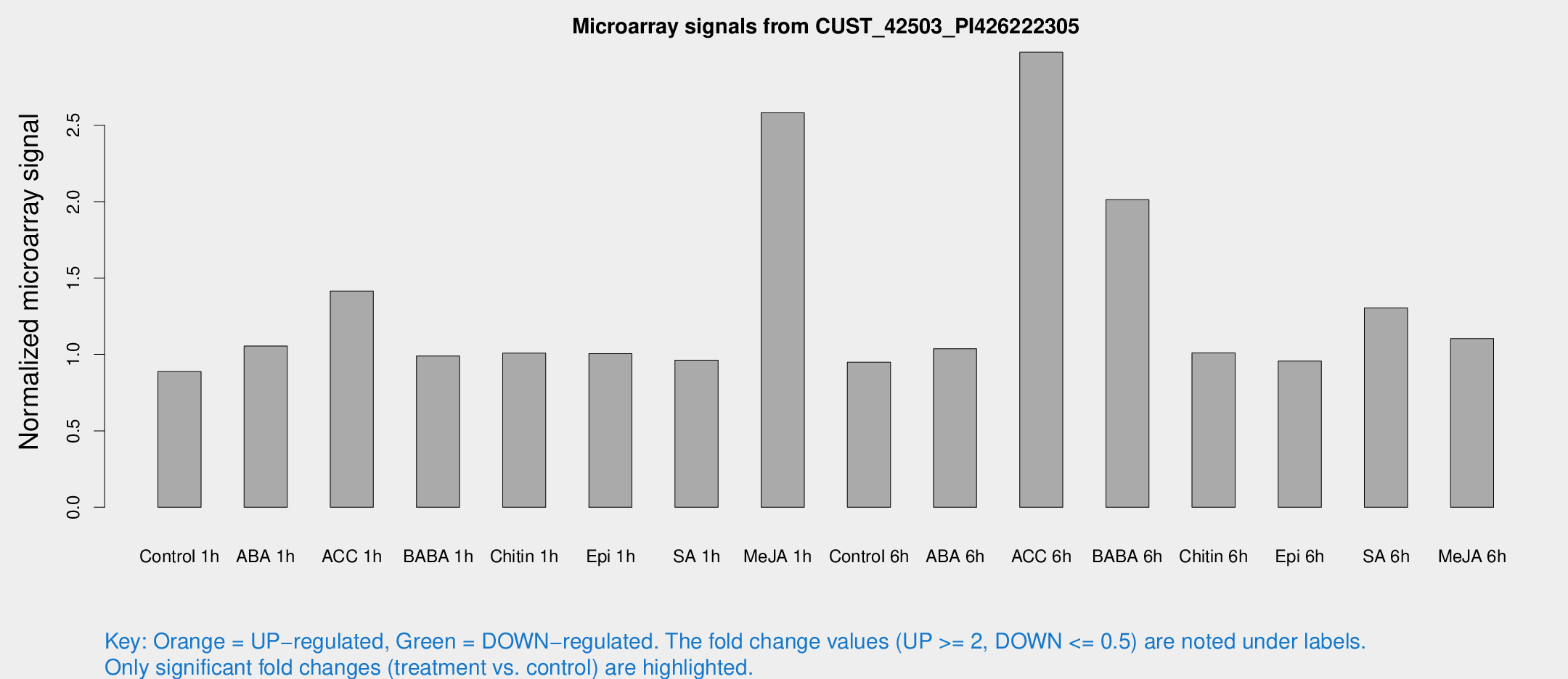
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.3296 | 3.67476 | 0.88745 | 0.514878 |
| ABA 1h | 6.68347 | 3.57144 | 1.05469 | 0.569939 |
| ACC 1h | 12.455 | 5.60851 | 1.41441 | 0.846369 |
| BABA 1h | 6.81914 | 3.96205 | 0.9894 | 0.572958 |
| Chitin 1h | 6.49048 | 3.77957 | 1.00866 | 0.58506 |
| Epi 1h | 6.22777 | 3.62854 | 1.00554 | 0.583742 |
| SA 1h | 7.17354 | 3.71747 | 0.962088 | 0.511276 |
| Me-JA 1h | 16.5646 | 4.60408 | 2.58068 | 0.904266 |
| Control 6h | 6.79967 | 3.94534 | 0.949979 | 0.550128 |
| ABA 6h | 8.0684 | 4.11482 | 1.03788 | 0.546953 |
| ACC 6h | 32.9692 | 14.0597 | 2.9773 | 2.15147 |
| BABA 6h | 18.7157 | 6.14699 | 2.01297 | 1.03494 |
| Chitin 6h | 7.67727 | 4.45422 | 1.00996 | 0.584829 |
| Epi 6h | 7.80345 | 4.52229 | 0.957031 | 0.549072 |
| SA 6h | 9.84192 | 4.19138 | 1.30377 | 0.633419 |
| Me-JA 6h | 8.07667 | 3.77218 | 1.10364 | 0.551899 |
Source Transcript PGSC0003DMT400079243 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
