Probe CUST_42226_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42226_PI426222305 | JHI_St_60k_v1 | DMT400006306 | CTCATGACAAGGTGGTTTCATTTCTTCTATTTCTCAAAAATAAGGTCTTCAAAGCTCCAA |
All Microarray Probes Designed to Gene DMG400002463
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42226_PI426222305 | JHI_St_60k_v1 | DMT400006306 | CTCATGACAAGGTGGTTTCATTTCTTCTATTTCTCAAAAATAAGGTCTTCAAAGCTCCAA |
Microarray Signals from CUST_42226_PI426222305
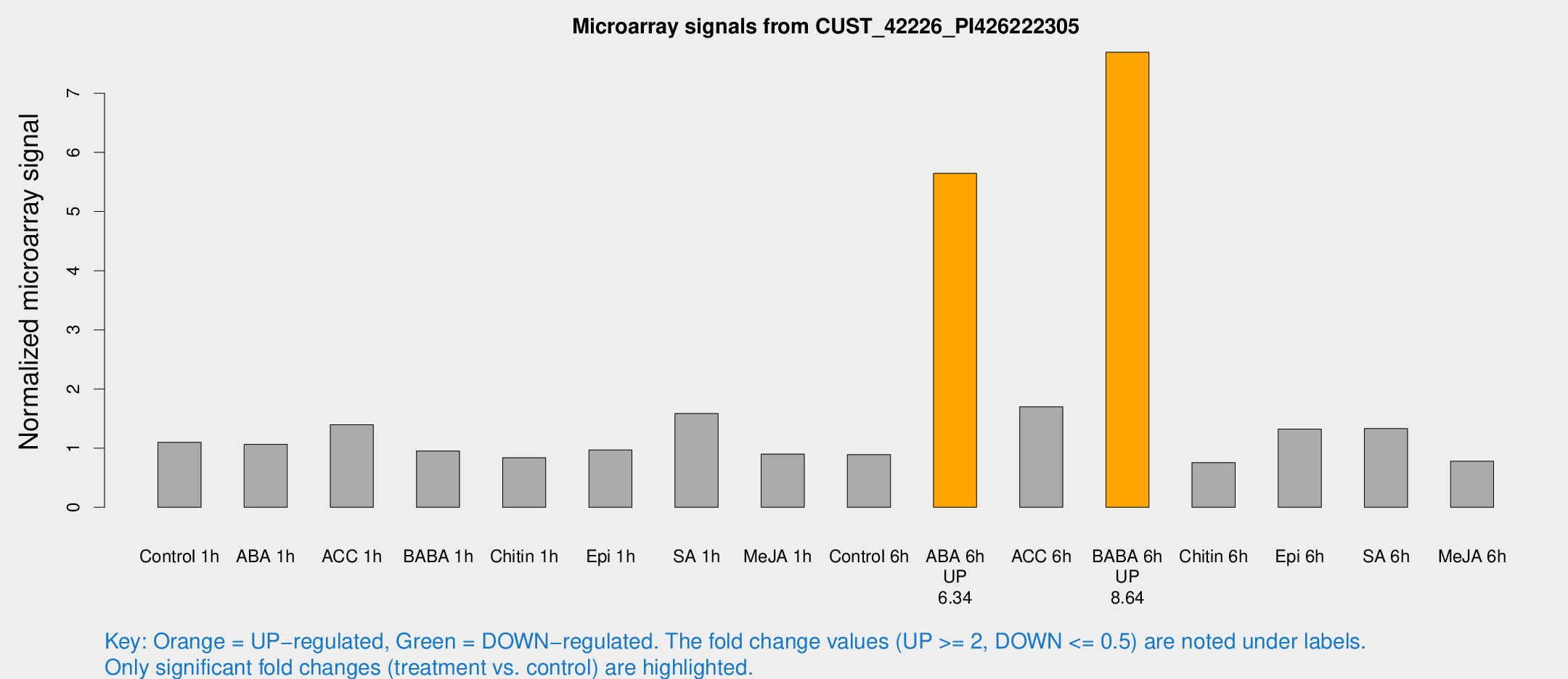
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9.48327 | 4.48549 | 1.09989 | 0.497987 |
| ABA 1h | 7.17052 | 2.8298 | 1.06381 | 0.486934 |
| ACC 1h | 10.7808 | 3.31307 | 1.39547 | 0.512836 |
| BABA 1h | 6.912 | 3.11888 | 0.951591 | 0.473498 |
| Chitin 1h | 5.36236 | 2.93222 | 0.837309 | 0.456305 |
| Epi 1h | 6.09325 | 2.92784 | 0.970433 | 0.489799 |
| SA 1h | 13.6522 | 5.00596 | 1.58454 | 0.640074 |
| Me-JA 1h | 5.12039 | 2.97611 | 0.899738 | 0.508851 |
| Control 6h | 6.58689 | 3.01549 | 0.890096 | 0.441878 |
| ABA 6h | 45.3947 | 11.0384 | 5.64331 | 1.12594 |
| ACC 6h | 14.5737 | 3.7303 | 1.70011 | 0.463087 |
| BABA 6h | 63.7046 | 11.5445 | 7.69255 | 1.69628 |
| Chitin 6h | 5.72335 | 3.32346 | 0.755193 | 0.438411 |
| Epi 6h | 11.1299 | 3.52522 | 1.32143 | 0.493981 |
| SA 6h | 10.3789 | 3.32767 | 1.33153 | 0.524161 |
| Me-JA 6h | 5.52375 | 2.979 | 0.77882 | 0.421785 |
Source Transcript PGSC0003DMT400006306 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G74590.1 | +1 | 7e-16 | 71 | 38/117 (32%) | glutathione S-transferase TAU 10 | chr1:28023887-28024666 REVERSE LENGTH=232 |
