Probe CUST_42212_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42212_PI426222305 | JHI_St_60k_v1 | DMT400038236 | GTGTGTCAGTGTGCTTACATTTCTCATGTTAGCAGTCATGTTGTTTCATTTTGTATTCAA |
All Microarray Probes Designed to Gene DMG400014753
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42212_PI426222305 | JHI_St_60k_v1 | DMT400038236 | GTGTGTCAGTGTGCTTACATTTCTCATGTTAGCAGTCATGTTGTTTCATTTTGTATTCAA |
Microarray Signals from CUST_42212_PI426222305
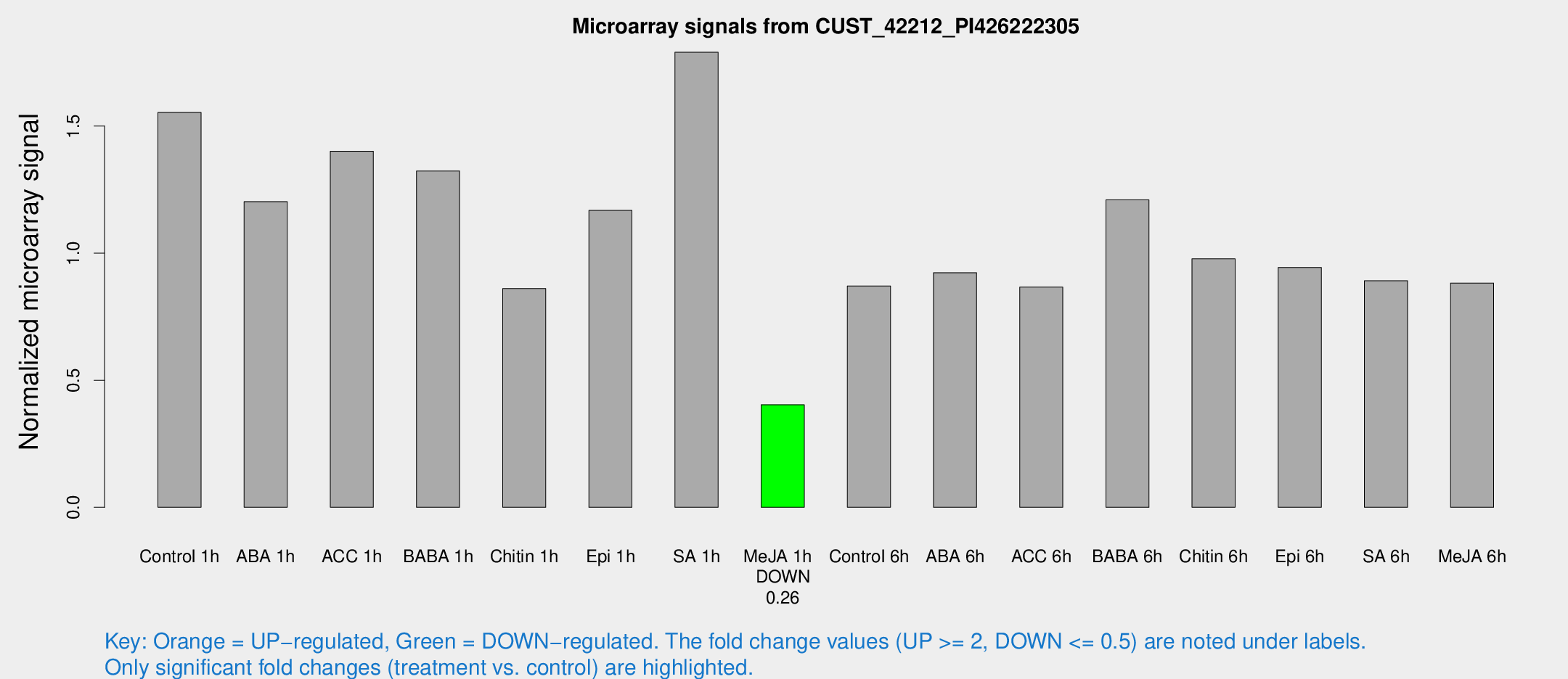
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2444.64 | 333.482 | 1.55403 | 0.107348 |
| ABA 1h | 1695.45 | 276.473 | 1.20281 | 0.164127 |
| ACC 1h | 2440.36 | 642.854 | 1.40092 | 0.363335 |
| BABA 1h | 2086.4 | 450.238 | 1.32339 | 0.193256 |
| Chitin 1h | 1227.66 | 203.6 | 0.860937 | 0.0749441 |
| Epi 1h | 1582.29 | 170.5 | 1.1683 | 0.123223 |
| SA 1h | 2905.88 | 443.313 | 1.79081 | 0.163208 |
| Me-JA 1h | 524.99 | 90.978 | 0.40348 | 0.0361793 |
| Control 6h | 1436.04 | 321.511 | 0.871081 | 0.147345 |
| ABA 6h | 1652.45 | 490.005 | 0.92323 | 0.225493 |
| ACC 6h | 1543.46 | 103.546 | 0.866549 | 0.0710802 |
| BABA 6h | 2149.97 | 353.809 | 1.20983 | 0.152668 |
| Chitin 6h | 1618.72 | 122.695 | 0.9779 | 0.084653 |
| Epi 6h | 1650.83 | 102.2 | 0.943737 | 0.0545387 |
| SA 6h | 1435.03 | 298.127 | 0.891422 | 0.10697 |
| Me-JA 6h | 1570.66 | 586.336 | 0.882593 | 0.284079 |
Source Transcript PGSC0003DMT400038236 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G34930.1 | +3 | 2e-110 | 366 | 304/946 (32%) | disease resistance family protein / LRR family protein | chr2:14737169-14739886 REVERSE LENGTH=905 |
