Probe CUST_42200_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42200_PI426222305 | JHI_St_60k_v1 | DMT400038220 | GATCGTAGTTGGGAAATGTTACTTGCAAAGTTACCAATATGTTGAGAGATATGGTTTAGG |
All Microarray Probes Designed to Gene DMG400014742
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42200_PI426222305 | JHI_St_60k_v1 | DMT400038220 | GATCGTAGTTGGGAAATGTTACTTGCAAAGTTACCAATATGTTGAGAGATATGGTTTAGG |
Microarray Signals from CUST_42200_PI426222305
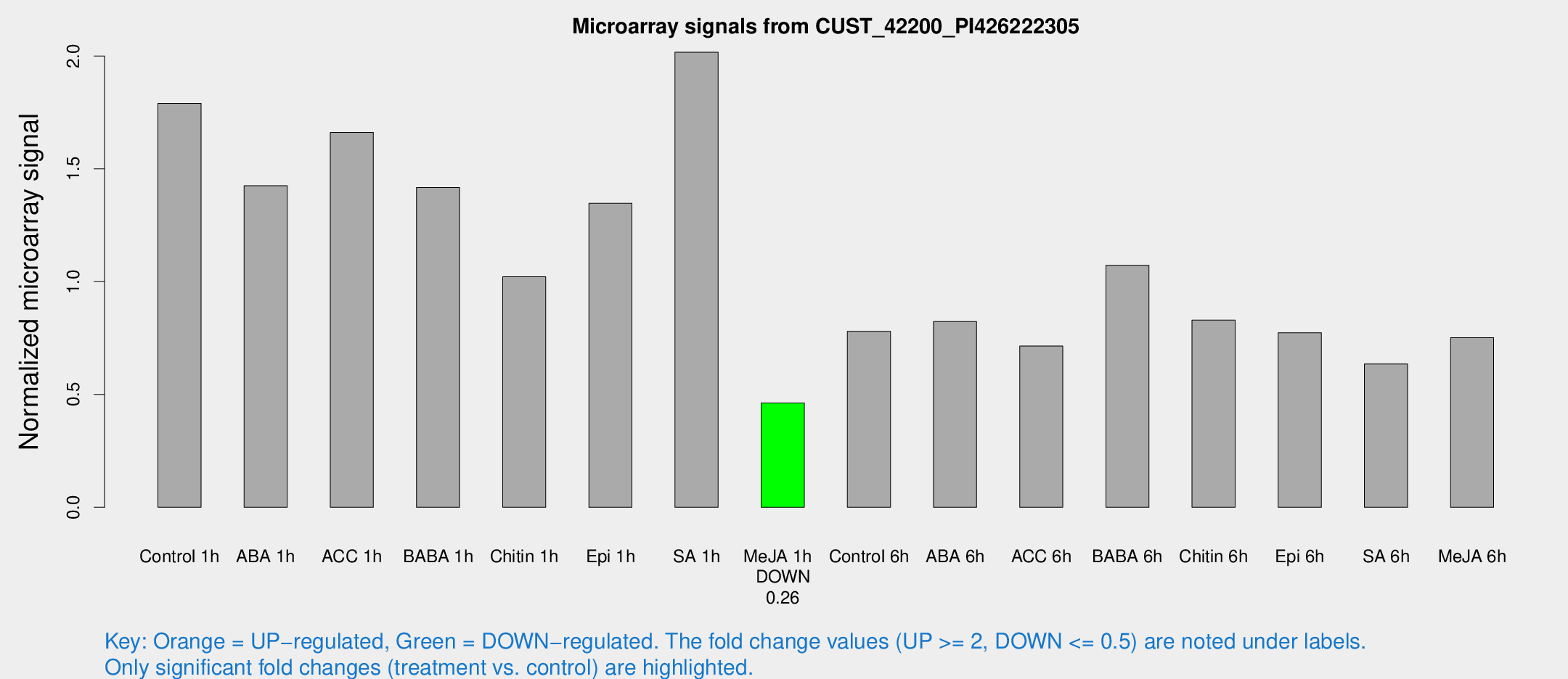
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1407.71 | 165.436 | 1.78979 | 0.103439 |
| ABA 1h | 992.173 | 111.592 | 1.42513 | 0.116983 |
| ACC 1h | 1457.48 | 389.922 | 1.66164 | 0.440526 |
| BABA 1h | 1123.91 | 243.024 | 1.41747 | 0.210928 |
| Chitin 1h | 725.086 | 94.8882 | 1.02143 | 0.0592114 |
| Epi 1h | 913.478 | 84.8132 | 1.34706 | 0.109663 |
| SA 1h | 1642.45 | 246.686 | 2.01677 | 0.204752 |
| Me-JA 1h | 302.878 | 55.2727 | 0.462682 | 0.0432441 |
| Control 6h | 662.823 | 174.341 | 0.779983 | 0.176005 |
| ABA 6h | 728.755 | 196.382 | 0.823383 | 0.175228 |
| ACC 6h | 638.97 | 40.167 | 0.714719 | 0.0561023 |
| BABA 6h | 942.897 | 103.022 | 1.07279 | 0.0944729 |
| Chitin 6h | 686.484 | 39.9317 | 0.829393 | 0.0481697 |
| Epi 6h | 677.427 | 39.3813 | 0.77356 | 0.0454979 |
| SA 6h | 531.355 | 137.071 | 0.635232 | 0.124801 |
| Me-JA 6h | 619.427 | 151.679 | 0.751956 | 0.130959 |
Source Transcript PGSC0003DMT400038220 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G74190.1 | +3 | 9e-24 | 100 | 72/178 (40%) | receptor like protein 15 | chr1:27902590-27906158 REVERSE LENGTH=965 |
