Probe CUST_41930_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41930_PI426222305 | JHI_St_60k_v1 | DMT400026490 | TGTGTAATCCATCTATGTTGTGCTTTGTCGTCTCCGAATTGAAGTTTGGTTTGTTATTTC |
All Microarray Probes Designed to Gene DMG400010222
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41930_PI426222305 | JHI_St_60k_v1 | DMT400026490 | TGTGTAATCCATCTATGTTGTGCTTTGTCGTCTCCGAATTGAAGTTTGGTTTGTTATTTC |
Microarray Signals from CUST_41930_PI426222305
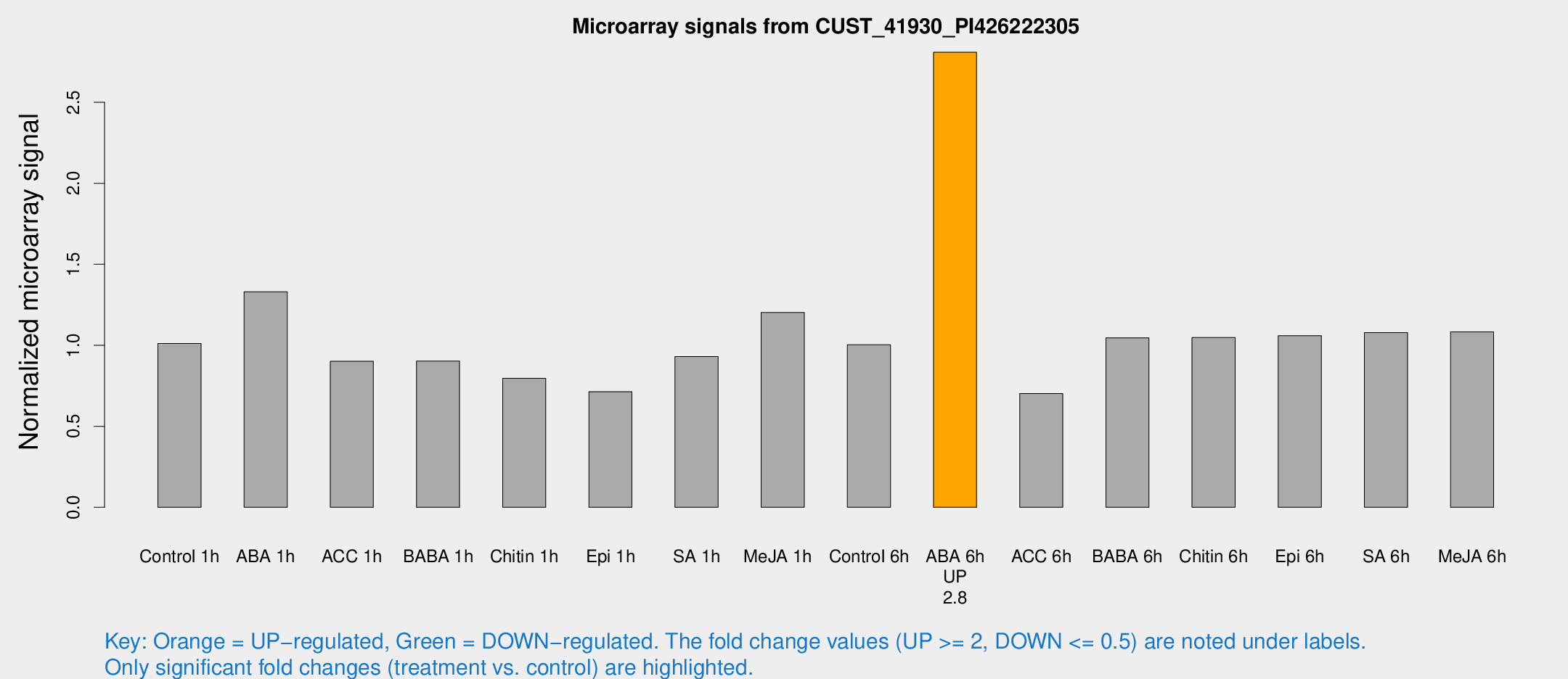
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 504.21 | 152.151 | 1.01101 | 0.24492 |
| ABA 1h | 556.383 | 99.9859 | 1.33054 | 0.263824 |
| ACC 1h | 473.22 | 140.553 | 0.901595 | 0.26143 |
| BABA 1h | 403.768 | 44.5569 | 0.902334 | 0.119527 |
| Chitin 1h | 353.692 | 84.6982 | 0.796012 | 0.184217 |
| Epi 1h | 300.206 | 69.3667 | 0.714012 | 0.162672 |
| SA 1h | 469.579 | 119.09 | 0.930173 | 0.303452 |
| Me-JA 1h | 456.749 | 54.8057 | 1.20251 | 0.148259 |
| Control 6h | 497.135 | 119.391 | 1.00327 | 0.201331 |
| ABA 6h | 1375.13 | 79.7222 | 2.80928 | 0.249548 |
| ACC 6h | 375.547 | 46.109 | 0.702007 | 0.0594457 |
| BABA 6h | 563.369 | 119.633 | 1.04635 | 0.270992 |
| Chitin 6h | 523.893 | 81.1972 | 1.04786 | 0.153556 |
| Epi 6h | 554.135 | 58.7635 | 1.05978 | 0.0615526 |
| SA 6h | 522.46 | 130.478 | 1.07794 | 0.182139 |
| Me-JA 6h | 496.416 | 28.9055 | 1.08311 | 0.138369 |
Source Transcript PGSC0003DMT400026490 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G18550.1 | +1 | 1e-139 | 411 | 227/401 (57%) | alpha/beta-Hydrolases superfamily protein | chr4:10225006-10226862 REVERSE LENGTH=419 |
