Probe CUST_41919_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41919_PI426222305 | JHI_St_60k_v1 | DMT400026480 | ATGGTTCAACAGGAAAATGGATCTTGGCTTCTGTTGGATCGCGACGAGTATGACTTCTGA |
All Microarray Probes Designed to Gene DMG400010219
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41919_PI426222305 | JHI_St_60k_v1 | DMT400026480 | ATGGTTCAACAGGAAAATGGATCTTGGCTTCTGTTGGATCGCGACGAGTATGACTTCTGA |
Microarray Signals from CUST_41919_PI426222305
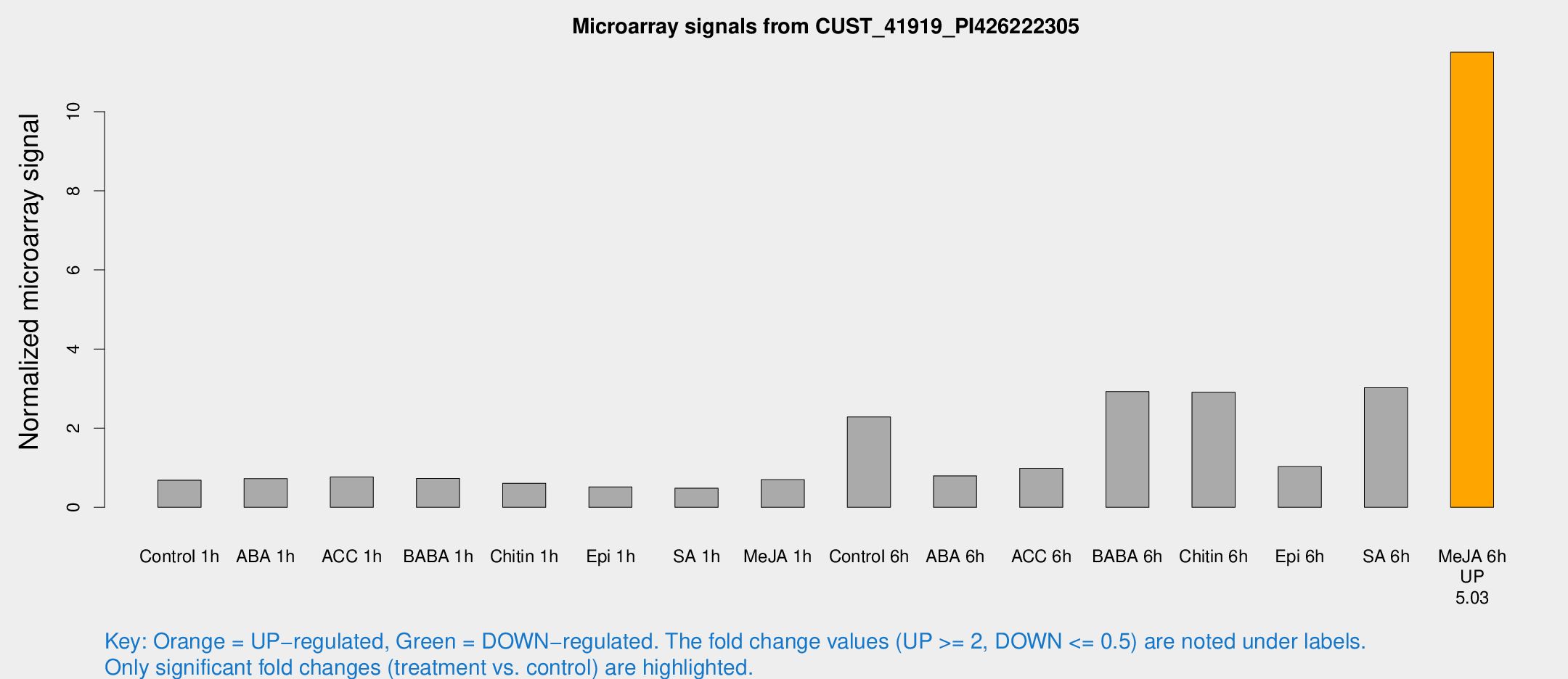
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 14.2466 | 4.27632 | 0.684879 | 0.289699 |
| ABA 1h | 12.7288 | 3.20489 | 0.724797 | 0.287316 |
| ACC 1h | 16.084 | 5.04006 | 0.766059 | 0.306086 |
| BABA 1h | 15.5196 | 6.07124 | 0.73075 | 0.284975 |
| Chitin 1h | 11.1267 | 3.26392 | 0.60857 | 0.221856 |
| Epi 1h | 9.98523 | 4.60962 | 0.517006 | 0.238484 |
| SA 1h | 10.5583 | 4.17949 | 0.483746 | 0.201965 |
| Me-JA 1h | 11.2061 | 3.4 | 0.696981 | 0.246331 |
| Control 6h | 51.0761 | 23.5722 | 2.28477 | 0.930465 |
| ABA 6h | 16.7476 | 4.84432 | 0.796218 | 0.2696 |
| ACC 6h | 25.8023 | 10.643 | 0.986581 | 0.558055 |
| BABA 6h | 67.649 | 21.6923 | 2.92717 | 1.24719 |
| Chitin 6h | 60.6419 | 13.8759 | 2.90841 | 0.733112 |
| Epi 6h | 26.6139 | 12.1467 | 1.02802 | 0.474557 |
| SA 6h | 66.6812 | 23.4077 | 3.02384 | 1.24725 |
| Me-JA 6h | 216.276 | 34.4272 | 11.5027 | 1.30446 |
Source Transcript PGSC0003DMT400026480 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G18550.1 | +1 | 1e-108 | 327 | 175/337 (52%) | alpha/beta-Hydrolases superfamily protein | chr4:10225006-10226862 REVERSE LENGTH=419 |
