Probe CUST_41916_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41916_PI426222305 | JHI_St_60k_v1 | DMT400026532 | TAAAATTAAATATGCTCATTAGGGGGTTCTGCAATTCTCTGTTTGCTCTCTTTTTGTCTC |
All Microarray Probes Designed to Gene DMG400010243
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41916_PI426222305 | JHI_St_60k_v1 | DMT400026532 | TAAAATTAAATATGCTCATTAGGGGGTTCTGCAATTCTCTGTTTGCTCTCTTTTTGTCTC |
Microarray Signals from CUST_41916_PI426222305
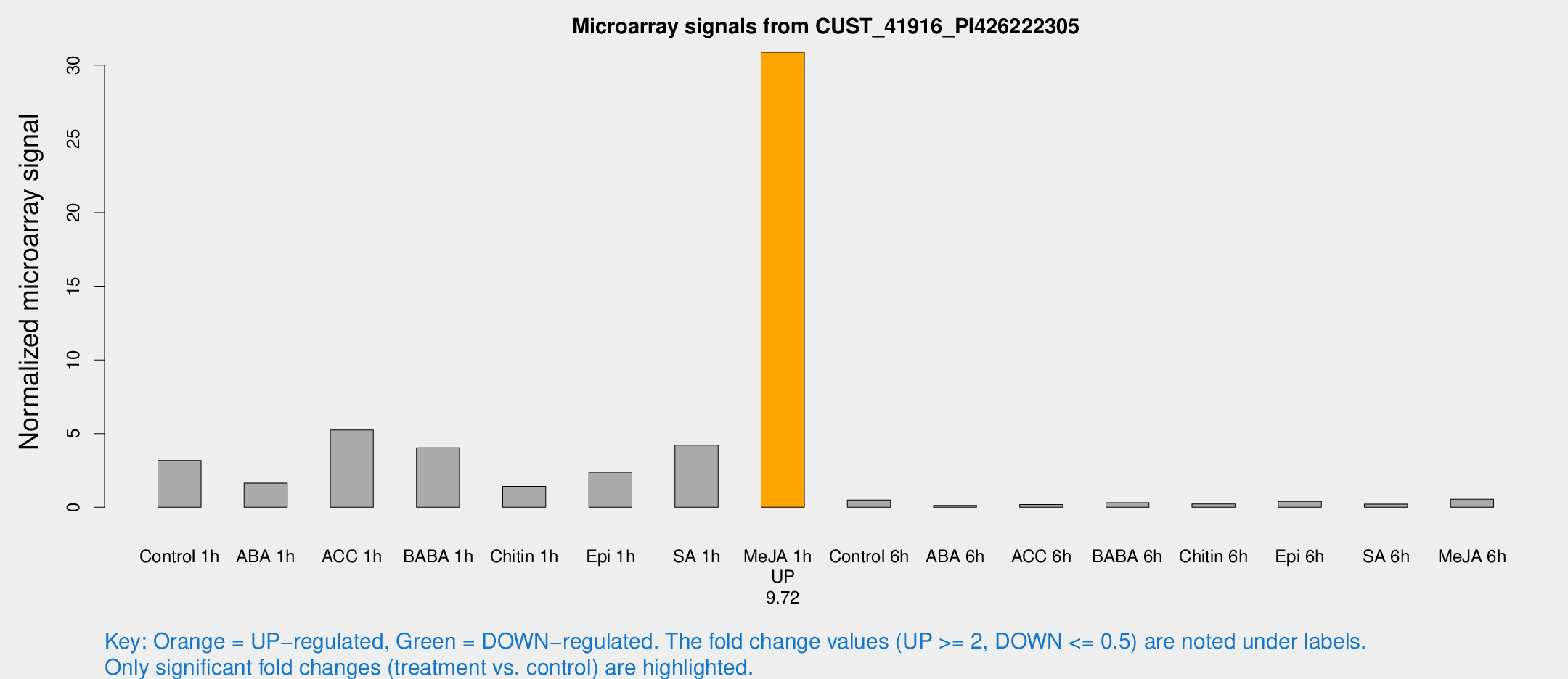
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 197.713 | 54.5824 | 3.17679 | 0.756983 |
| ABA 1h | 90.7766 | 25.9949 | 1.64543 | 0.341184 |
| ACC 1h | 385.464 | 171.081 | 5.24972 | 3.03434 |
| BABA 1h | 230.134 | 32.8173 | 4.03485 | 0.919176 |
| Chitin 1h | 83.1054 | 28.7782 | 1.41844 | 0.492988 |
| Epi 1h | 123.388 | 23.4461 | 2.38278 | 0.480408 |
| SA 1h | 436.493 | 300.302 | 4.20925 | 5.66823 |
| Me-JA 1h | 1462.69 | 84.7836 | 30.8777 | 1.78442 |
| Control 6h | 34.2207 | 13.4926 | 0.498235 | 0.180372 |
| ABA 6h | 7.97666 | 4.0541 | 0.127444 | 0.0666828 |
| ACC 6h | 12.6244 | 4.80288 | 0.185 | 0.0750955 |
| BABA 6h | 20.57 | 4.34762 | 0.309672 | 0.0721424 |
| Chitin 6h | 15.4205 | 4.29926 | 0.227592 | 0.0814706 |
| Epi 6h | 33.3348 | 14.7928 | 0.392425 | 0.342018 |
| SA 6h | 17.5425 | 10.4343 | 0.218015 | 0.136599 |
| Me-JA 6h | 31.845 | 4.7899 | 0.540162 | 0.115101 |
Source Transcript PGSC0003DMT400026532 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G06330.1 | +3 | 8e-30 | 110 | 60/123 (49%) | Heavy metal transport/detoxification superfamily protein | chr1:1931671-1932266 REVERSE LENGTH=159 |
