Probe CUST_41912_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41912_PI426222305 | JHI_St_60k_v1 | DMT400026550 | GGACACGTTAAAAGATGAATATGGTGTACCTATTTCATGGTGGGTTGTAGAGAACAAAGG |
All Microarray Probes Designed to Gene DMG400010250
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41912_PI426222305 | JHI_St_60k_v1 | DMT400026550 | GGACACGTTAAAAGATGAATATGGTGTACCTATTTCATGGTGGGTTGTAGAGAACAAAGG |
Microarray Signals from CUST_41912_PI426222305
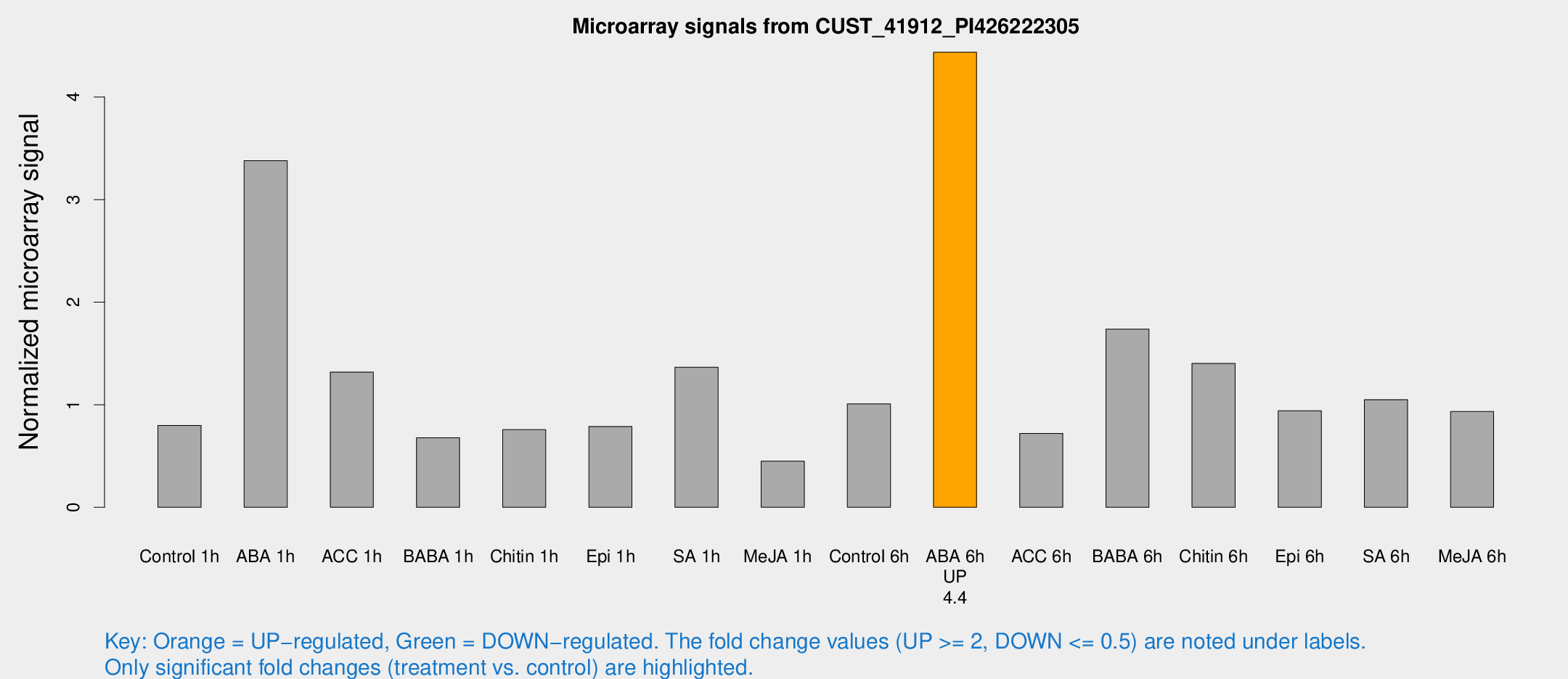
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 20.8647 | 6.60601 | 0.798153 | 0.201103 |
| ABA 1h | 74.5486 | 16.177 | 3.37953 | 0.717429 |
| ACC 1h | 34.281 | 8.77066 | 1.31788 | 0.450954 |
| BABA 1h | 22.1891 | 12.3738 | 0.678059 | 0.444233 |
| Chitin 1h | 17.5527 | 5.02567 | 0.757274 | 0.15982 |
| Epi 1h | 19.5478 | 7.6675 | 0.787903 | 0.385151 |
| SA 1h | 35.4409 | 8.16799 | 1.36585 | 0.399974 |
| Me-JA 1h | 9.63944 | 3.2229 | 0.450064 | 0.190216 |
| Control 6h | 29.6286 | 14.0417 | 1.00798 | 0.432878 |
| ABA 6h | 123.993 | 40.3648 | 4.43663 | 1.15875 |
| ACC 6h | 23.9438 | 8.33091 | 0.720921 | 0.354433 |
| BABA 6h | 46.5202 | 4.44345 | 1.7374 | 0.165725 |
| Chitin 6h | 38.184 | 9.36931 | 1.40262 | 0.352928 |
| Epi 6h | 25.3832 | 3.96715 | 0.94125 | 0.145654 |
| SA 6h | 28.1087 | 8.31899 | 1.04983 | 0.292773 |
| Me-JA 6h | 24.6099 | 7.74284 | 0.933334 | 0.235246 |
Source Transcript PGSC0003DMT400026550 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G18550.1 | +1 | 2e-162 | 467 | 236/397 (59%) | alpha/beta-Hydrolases superfamily protein | chr4:10225006-10226862 REVERSE LENGTH=419 |
