Probe CUST_41910_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41910_PI426222305 | JHI_St_60k_v1 | DMT400026504 | GCCCTTGCCTTGAGATATGAATGAATGACATTTGATTCTATATGACATTCTGGTATAGTA |
All Microarray Probes Designed to Gene DMG400010231
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41910_PI426222305 | JHI_St_60k_v1 | DMT400026504 | GCCCTTGCCTTGAGATATGAATGAATGACATTTGATTCTATATGACATTCTGGTATAGTA |
Microarray Signals from CUST_41910_PI426222305
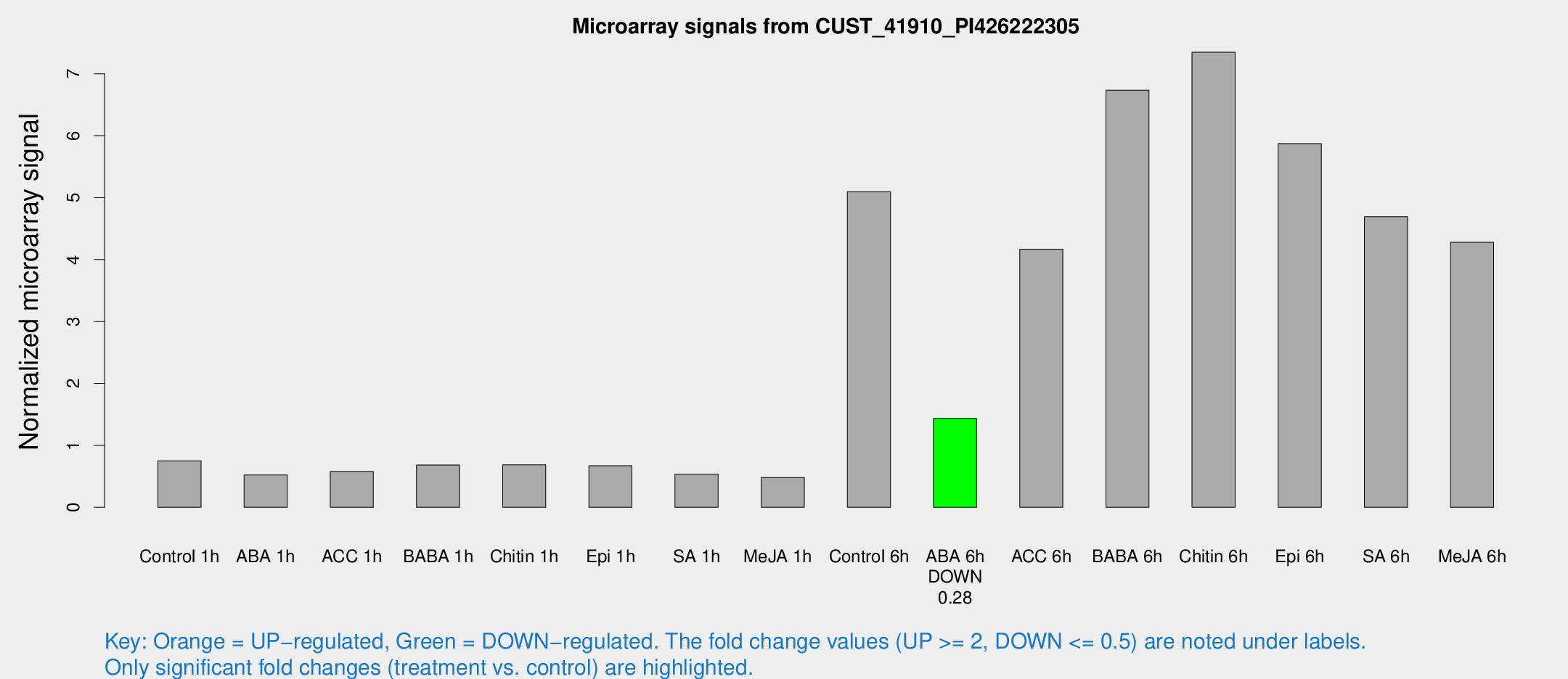
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 159.591 | 18.7212 | 0.750467 | 0.0459991 |
| ABA 1h | 98.6796 | 13.3728 | 0.521326 | 0.0671177 |
| ACC 1h | 146.225 | 50.3932 | 0.578613 | 0.221775 |
| BABA 1h | 139.641 | 15.4418 | 0.682105 | 0.149815 |
| Chitin 1h | 134.651 | 24.7953 | 0.685682 | 0.106632 |
| Epi 1h | 121.793 | 7.69597 | 0.670303 | 0.042366 |
| SA 1h | 119.32 | 21.7168 | 0.532556 | 0.141219 |
| Me-JA 1h | 83.0453 | 7.09968 | 0.481069 | 0.0850861 |
| Control 6h | 1136.14 | 265.438 | 5.09729 | 0.906986 |
| ABA 6h | 334.672 | 64.8522 | 1.43642 | 0.345928 |
| ACC 6h | 1025.83 | 158.44 | 4.16692 | 0.241108 |
| BABA 6h | 1610.25 | 207.935 | 6.73768 | 0.72555 |
| Chitin 6h | 1648.7 | 103.266 | 7.34851 | 0.424608 |
| Epi 6h | 1395.47 | 84.0456 | 5.87402 | 0.807674 |
| SA 6h | 992.46 | 127.871 | 4.69295 | 0.271527 |
| Me-JA 6h | 993.786 | 311.493 | 4.27911 | 1.09783 |
Source Transcript PGSC0003DMT400026504 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G29240.1 | +2 | 6e-33 | 135 | 162/606 (27%) | Protein of unknown function (DUF688) | chr1:10217023-10218924 REVERSE LENGTH=577 |
