Probe CUST_41904_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41904_PI426222305 | JHI_St_60k_v1 | DMT400026489 | CTTTCGTGTAATGTTTGAATAAAAGGTTTTCGAGGGCTGTTTGGAAAAATCACATCCTAT |
All Microarray Probes Designed to Gene DMG400010221
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41904_PI426222305 | JHI_St_60k_v1 | DMT400026489 | CTTTCGTGTAATGTTTGAATAAAAGGTTTTCGAGGGCTGTTTGGAAAAATCACATCCTAT |
Microarray Signals from CUST_41904_PI426222305
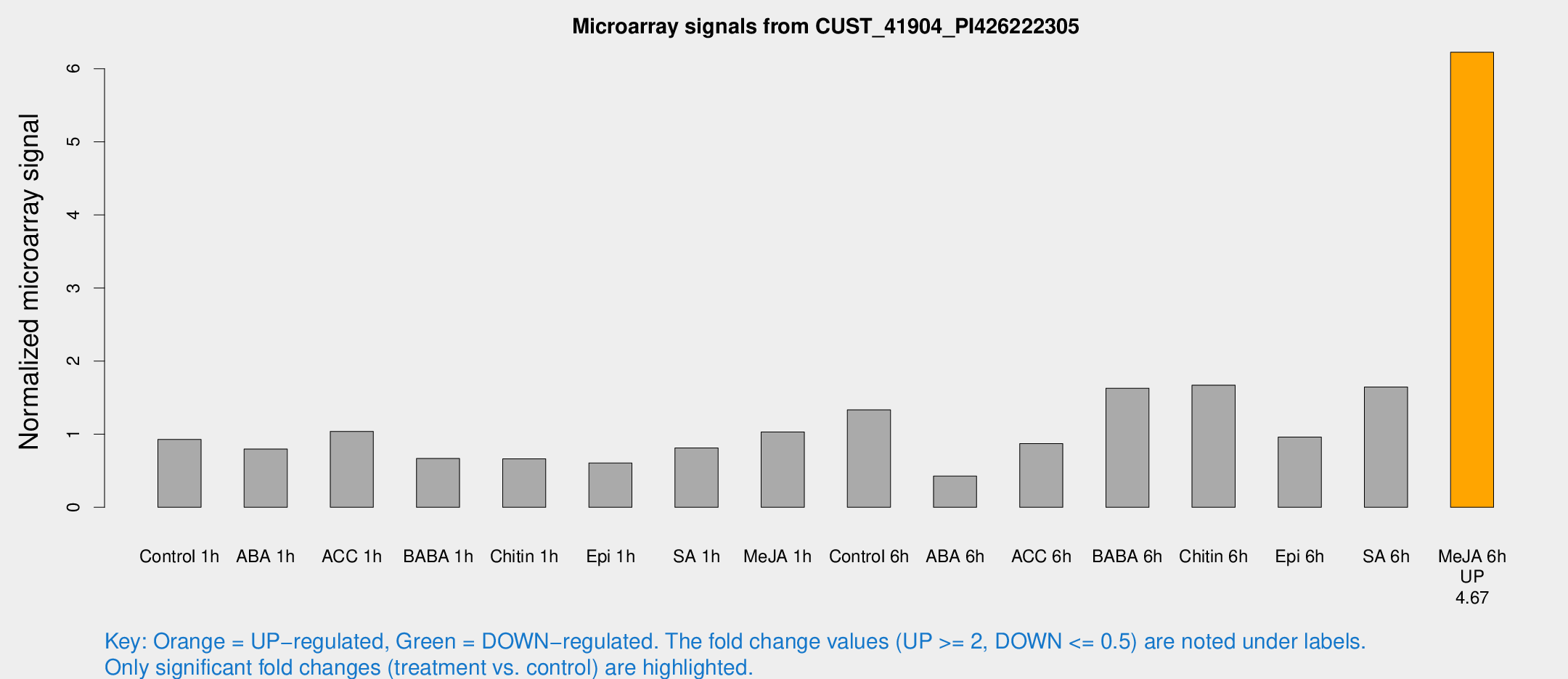
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 874.308 | 58.9081 | 0.928709 | 0.10027 |
| ABA 1h | 668.048 | 71.323 | 0.796794 | 0.0722354 |
| ACC 1h | 1027.05 | 165.625 | 1.03894 | 0.0981357 |
| BABA 1h | 611.779 | 72.3946 | 0.668069 | 0.151266 |
| Chitin 1h | 613.964 | 198.503 | 0.66322 | 0.219804 |
| Epi 1h | 500.393 | 66.9193 | 0.605835 | 0.072178 |
| SA 1h | 850.752 | 218.694 | 0.812571 | 0.319029 |
| Me-JA 1h | 830.639 | 197.675 | 1.03112 | 0.266943 |
| Control 6h | 1482.44 | 497.297 | 1.3326 | 0.521793 |
| ABA 6h | 471.754 | 161.092 | 0.426328 | 0.170688 |
| ACC 6h | 992.917 | 224.237 | 0.871664 | 0.135769 |
| BABA 6h | 1798.17 | 416.812 | 1.62912 | 0.439211 |
| Chitin 6h | 1668.24 | 96.4513 | 1.67171 | 0.0966071 |
| Epi 6h | 1025.3 | 96.5198 | 0.961411 | 0.0582332 |
| SA 6h | 1698.07 | 513.463 | 1.64497 | 0.402167 |
| Me-JA 6h | 6148.4 | 1450.97 | 6.22546 | 1.29673 |
Source Transcript PGSC0003DMT400026489 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G18550.1 | +2 | 1e-148 | 435 | 217/395 (55%) | alpha/beta-Hydrolases superfamily protein | chr4:10225006-10226862 REVERSE LENGTH=419 |
