Probe CUST_41896_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41896_PI426222305 | JHI_St_60k_v1 | DMT400026502 | GGGCCAAGCGATATTCAACACTCAGATACGGTTATTTGGTTCAATATATATACATAATAC |
All Microarray Probes Designed to Gene DMG400010229
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41883_PI426222305 | JHI_St_60k_v1 | DMT400026501 | GGGCCAAGCGATATTCAACACTCAGATACGGTTATTTGGTTCAATATATATACATAATAC |
| CUST_41896_PI426222305 | JHI_St_60k_v1 | DMT400026502 | GGGCCAAGCGATATTCAACACTCAGATACGGTTATTTGGTTCAATATATATACATAATAC |
Microarray Signals from CUST_41896_PI426222305
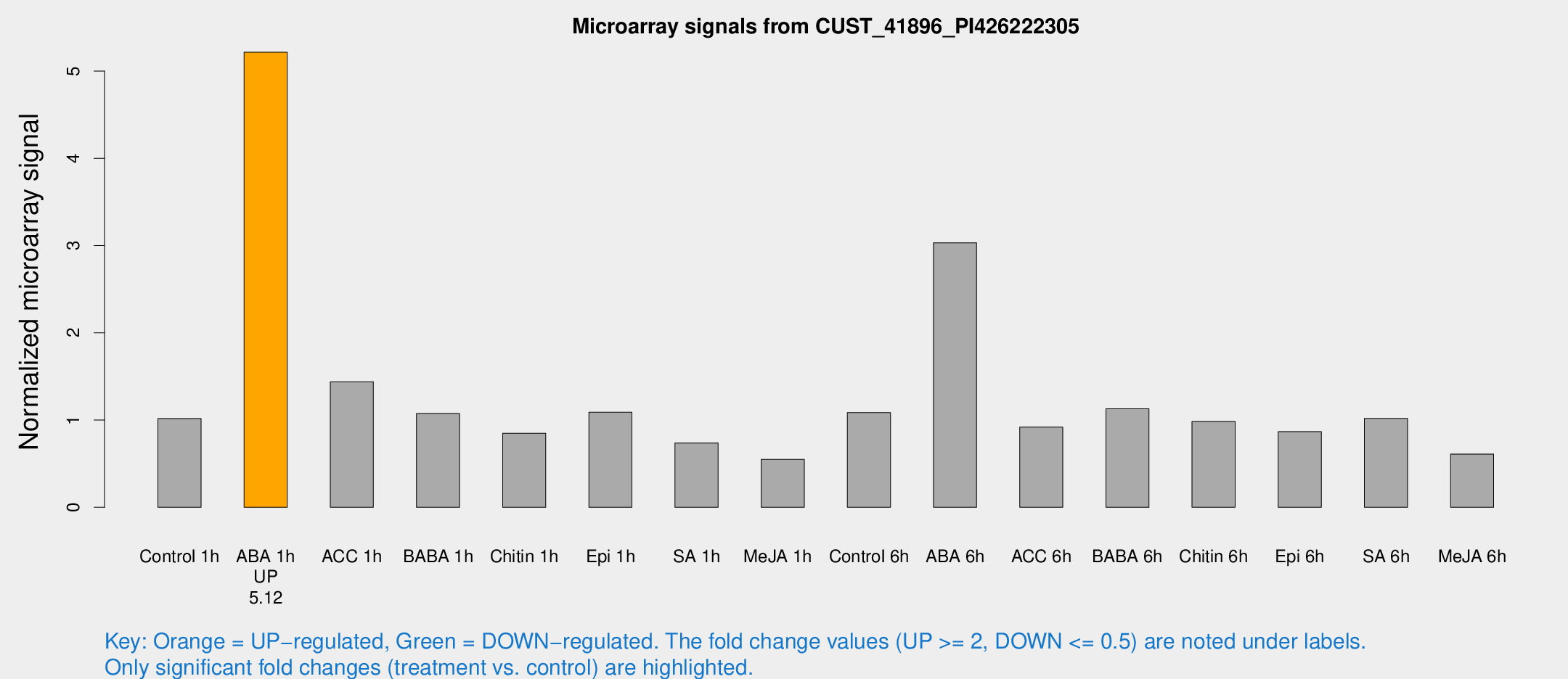
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 169.008 | 19.5253 | 1.01804 | 0.150523 |
| ABA 1h | 755.991 | 43.8455 | 5.21648 | 0.404415 |
| ACC 1h | 242.936 | 16.1998 | 1.43944 | 0.203194 |
| BABA 1h | 172.856 | 22.8917 | 1.07524 | 0.0669005 |
| Chitin 1h | 125.338 | 8.11204 | 0.849169 | 0.0665612 |
| Epi 1h | 156.382 | 18.186 | 1.08913 | 0.155333 |
| SA 1h | 124.811 | 10.6804 | 0.737208 | 0.0476815 |
| Me-JA 1h | 73.4032 | 5.98298 | 0.549302 | 0.0499495 |
| Control 6h | 206.22 | 66.2644 | 1.08504 | 0.376345 |
| ABA 6h | 545.073 | 98.08 | 3.03223 | 0.388261 |
| ACC 6h | 178.302 | 33.6709 | 0.918621 | 0.0578759 |
| BABA 6h | 206.961 | 12.6691 | 1.12935 | 0.0691195 |
| Chitin 6h | 171.636 | 10.7407 | 0.983717 | 0.0615435 |
| Epi 6h | 162.154 | 17.8416 | 0.867696 | 0.155475 |
| SA 6h | 165.168 | 10.3074 | 1.019 | 0.113443 |
| Me-JA 6h | 107.902 | 27.5908 | 0.610415 | 0.137686 |
Source Transcript PGSC0003DMT400026502 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G18550.1 | +2 | 1e-72 | 233 | 121/199 (61%) | alpha/beta-Hydrolases superfamily protein | chr4:10225006-10226862 REVERSE LENGTH=419 |
