Probe CUST_41890_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41890_PI426222305 | JHI_St_60k_v1 | DMT400026503 | CTGAATTATACTTCACTTAATGTCAACTGCAAGTCCTGTGTACCACATTTTTCTGTTCTA |
All Microarray Probes Designed to Gene DMG400010230
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41890_PI426222305 | JHI_St_60k_v1 | DMT400026503 | CTGAATTATACTTCACTTAATGTCAACTGCAAGTCCTGTGTACCACATTTTTCTGTTCTA |
Microarray Signals from CUST_41890_PI426222305
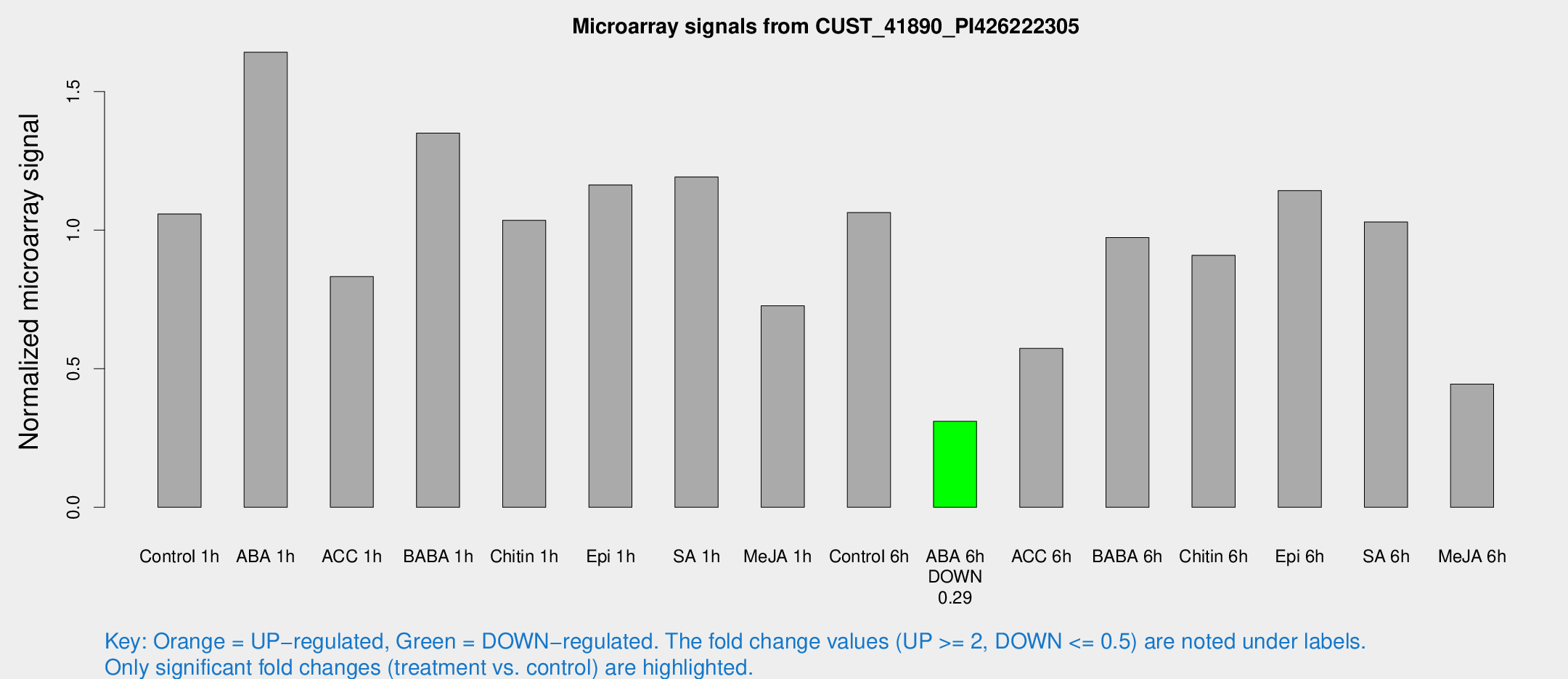
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 882.684 | 153.192 | 1.05859 | 0.110779 |
| ABA 1h | 1202.63 | 168.776 | 1.64217 | 0.158418 |
| ACC 1h | 766.308 | 208.509 | 0.832701 | 0.230601 |
| BABA 1h | 1105.39 | 220.151 | 1.35034 | 0.166331 |
| Chitin 1h | 791.065 | 173.309 | 1.0356 | 0.146297 |
| Epi 1h | 817.496 | 47.4054 | 1.16341 | 0.0672998 |
| SA 1h | 1082.52 | 333.223 | 1.19201 | 0.286419 |
| Me-JA 1h | 504.368 | 115.568 | 0.727142 | 0.112589 |
| Control 6h | 935.808 | 235.02 | 1.06339 | 0.224594 |
| ABA 6h | 270.184 | 27.9554 | 0.310283 | 0.0412861 |
| ACC 6h | 538.023 | 54.8762 | 0.573488 | 0.0333271 |
| BABA 6h | 891.958 | 89.4526 | 0.973521 | 0.0718923 |
| Chitin 6h | 788.67 | 59.7269 | 0.909381 | 0.0526498 |
| Epi 6h | 1061.22 | 141.262 | 1.14301 | 0.176946 |
| SA 6h | 831.14 | 71.8167 | 1.02945 | 0.059575 |
| Me-JA 6h | 420.607 | 162.191 | 0.444617 | 0.154984 |
Source Transcript PGSC0003DMT400026503 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G45840.2 | +2 | 0.0 | 608 | 342/724 (47%) | Leucine-rich repeat protein kinase family protein | chr5:18594080-18597221 REVERSE LENGTH=706 |
