Probe CUST_41883_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41883_PI426222305 | JHI_St_60k_v1 | DMT400026501 | GGGCCAAGCGATATTCAACACTCAGATACGGTTATTTGGTTCAATATATATACATAATAC |
All Microarray Probes Designed to Gene DMG400010229
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41896_PI426222305 | JHI_St_60k_v1 | DMT400026502 | GGGCCAAGCGATATTCAACACTCAGATACGGTTATTTGGTTCAATATATATACATAATAC |
| CUST_41883_PI426222305 | JHI_St_60k_v1 | DMT400026501 | GGGCCAAGCGATATTCAACACTCAGATACGGTTATTTGGTTCAATATATATACATAATAC |
Microarray Signals from CUST_41883_PI426222305
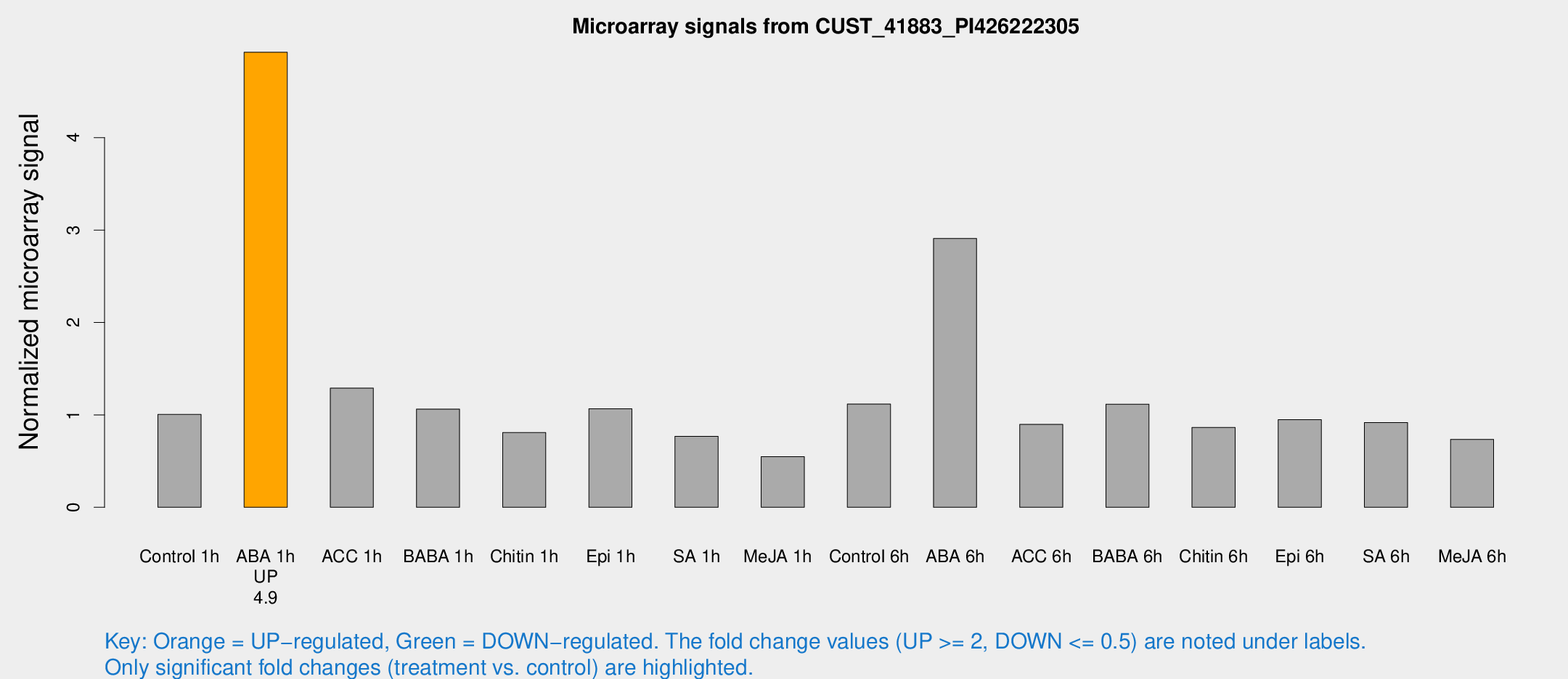
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 167.039 | 18.2119 | 1.00532 | 0.125869 |
| ABA 1h | 718.02 | 51.2842 | 4.92432 | 0.522756 |
| ACC 1h | 218.751 | 18.0172 | 1.28969 | 0.201196 |
| BABA 1h | 169.55 | 16.1576 | 1.06308 | 0.0857027 |
| Chitin 1h | 119.769 | 7.79242 | 0.808949 | 0.0524907 |
| Epi 1h | 152.142 | 10.8358 | 1.06625 | 0.0777477 |
| SA 1h | 129.821 | 8.32331 | 0.768578 | 0.0492078 |
| Me-JA 1h | 73.9815 | 6.08962 | 0.5489 | 0.089624 |
| Control 6h | 204.448 | 57.4928 | 1.11721 | 0.298553 |
| ABA 6h | 524.114 | 91.4069 | 2.90927 | 0.380354 |
| ACC 6h | 179.938 | 45.8047 | 0.897008 | 0.113466 |
| BABA 6h | 205.988 | 15.0114 | 1.11612 | 0.0681795 |
| Chitin 6h | 151.654 | 9.79078 | 0.864763 | 0.0664933 |
| Epi 6h | 179.632 | 26.5941 | 0.947744 | 0.21696 |
| SA 6h | 148.806 | 9.46737 | 0.916743 | 0.126557 |
| Me-JA 6h | 128.651 | 32.5635 | 0.735509 | 0.136096 |
Source Transcript PGSC0003DMT400026501 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G18550.1 | +3 | 6e-162 | 471 | 233/391 (60%) | alpha/beta-Hydrolases superfamily protein | chr4:10225006-10226862 REVERSE LENGTH=419 |
