Probe CUST_41597_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41597_PI426222305 | JHI_St_60k_v1 | DMT400095041 | AGTTTAACTTGGCTGGTCTTCATGCGGCAAATGGGTGTCCGATAGGAAAGCATTTGAAAC |
All Microarray Probes Designed to Gene DMG400044612
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41597_PI426222305 | JHI_St_60k_v1 | DMT400095041 | AGTTTAACTTGGCTGGTCTTCATGCGGCAAATGGGTGTCCGATAGGAAAGCATTTGAAAC |
Microarray Signals from CUST_41597_PI426222305
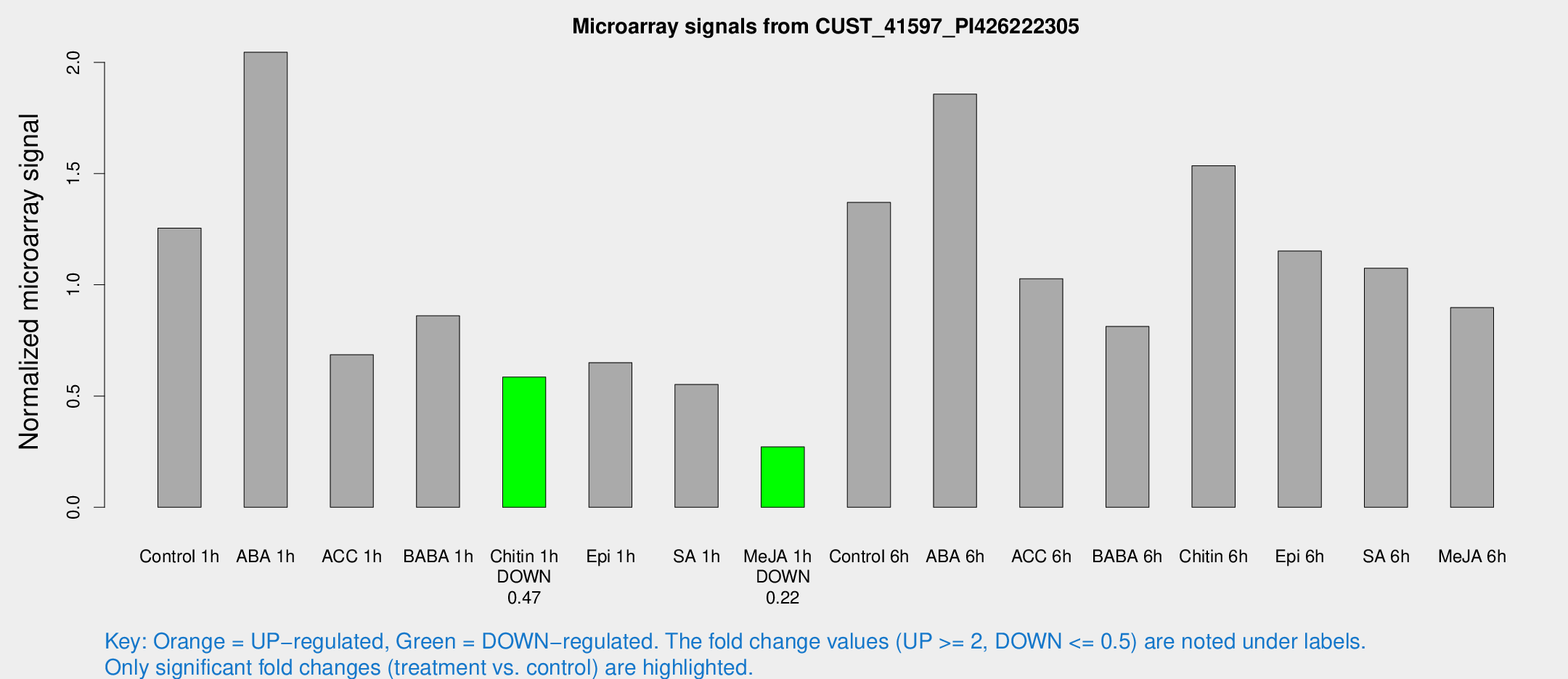
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 84.9573 | 9.51495 | 1.25436 | 0.106556 |
| ABA 1h | 123.626 | 18.4828 | 2.04532 | 0.443019 |
| ACC 1h | 47.4023 | 4.51243 | 0.685544 | 0.0672992 |
| BABA 1h | 56.9259 | 8.82117 | 0.860696 | 0.0743615 |
| Chitin 1h | 35.3049 | 3.80596 | 0.5854 | 0.062843 |
| Epi 1h | 39.5907 | 9.2965 | 0.650249 | 0.153534 |
| SA 1h | 39.3448 | 7.11137 | 0.551676 | 0.149891 |
| Me-JA 1h | 15.3821 | 3.47992 | 0.271406 | 0.0653485 |
| Control 6h | 97.1882 | 23.2817 | 1.3704 | 0.219335 |
| ABA 6h | 132.881 | 10.5656 | 1.85664 | 0.117716 |
| ACC 6h | 80.7999 | 12.959 | 1.0276 | 0.236796 |
| BABA 6h | 66.9823 | 21.4143 | 0.812566 | 0.248242 |
| Chitin 6h | 110.774 | 12.8974 | 1.53534 | 0.222835 |
| Epi 6h | 93.9067 | 25.3521 | 1.15161 | 0.406026 |
| SA 6h | 88.7647 | 38.0132 | 1.07425 | 0.86268 |
| Me-JA 6h | 63.2697 | 15.2796 | 0.89741 | 0.1891 |
Source Transcript PGSC0003DMT400095041 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
