Probe CUST_41534_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41534_PI426222305 | JHI_St_60k_v1 | DMT400021590 | GAAGATTATGTTCCAGAGTTGTTGGAGAATTTGCCTACAAATCATGTGTGGTATGACTAA |
All Microarray Probes Designed to Gene DMG400008372
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41534_PI426222305 | JHI_St_60k_v1 | DMT400021590 | GAAGATTATGTTCCAGAGTTGTTGGAGAATTTGCCTACAAATCATGTGTGGTATGACTAA |
| CUST_41491_PI426222305 | JHI_St_60k_v1 | DMT400021591 | TAGCCATGTCCAACTTGCTTTGATATTTTTGTATTTAGTGGCTAGTATTGCAACTTGGAG |
Microarray Signals from CUST_41534_PI426222305
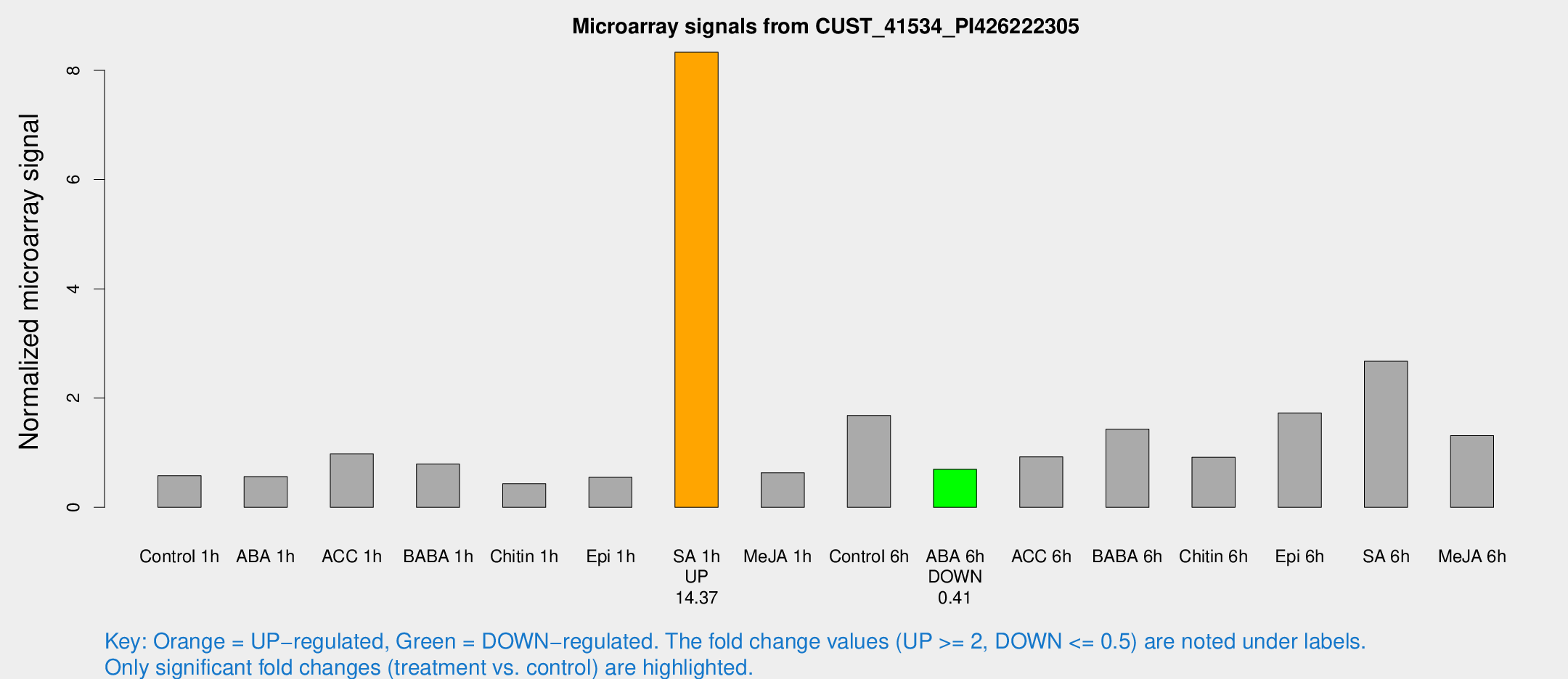
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1325.83 | 422.907 | 0.579678 | 0.144267 |
| ABA 1h | 1158.91 | 355.759 | 0.561858 | 0.188074 |
| ACC 1h | 2632.45 | 1098.59 | 0.975592 | 0.508078 |
| BABA 1h | 1778.48 | 522.987 | 0.790314 | 0.209676 |
| Chitin 1h | 829.302 | 125.91 | 0.431511 | 0.0619455 |
| Epi 1h | 1009.82 | 137.31 | 0.547626 | 0.0745787 |
| SA 1h | 19129.7 | 5241.76 | 8.33146 | 1.70688 |
| Me-JA 1h | 1158.5 | 329.859 | 0.631607 | 0.120526 |
| Control 6h | 3682.07 | 727.211 | 1.6818 | 0.213432 |
| ABA 6h | 1654.66 | 404.031 | 0.696554 | 0.156479 |
| ACC 6h | 2228.57 | 210.39 | 0.922752 | 0.206286 |
| BABA 6h | 3530.85 | 869.795 | 1.4306 | 0.295309 |
| Chitin 6h | 2062.85 | 219.714 | 0.917614 | 0.0965291 |
| Epi 6h | 4202.59 | 713.075 | 1.72758 | 0.224279 |
| SA 6h | 5672.38 | 871.765 | 2.67492 | 0.154448 |
| Me-JA 6h | 2974.84 | 928.421 | 1.31123 | 0.3012 |
Source Transcript PGSC0003DMT400021590 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
