Probe CUST_40406_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_40406_PI426222305 | JHI_St_60k_v1 | DMT400061609 | CACAATGTATATCTAGCCTTATTCGCTACAATGTTCTTCGCGTATCTTTGTACTTAAATG |
All Microarray Probes Designed to Gene DMG400023984
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_40396_PI426222305 | JHI_St_60k_v1 | DMT400061610 | TTCCCACCATGTATTTTCACAGTGAGTTTGATTCGATAGAAAATGACTGGAAAAATCATG |
| CUST_40429_PI426222305 | JHI_St_60k_v1 | DMT400061611 | AGTGTTTGGTGTTGTTTTTGGTTAGATTTTGATTAGGGTGGAGATGCATTATTAGGATGT |
| CUST_40406_PI426222305 | JHI_St_60k_v1 | DMT400061609 | CACAATGTATATCTAGCCTTATTCGCTACAATGTTCTTCGCGTATCTTTGTACTTAAATG |
Microarray Signals from CUST_40406_PI426222305
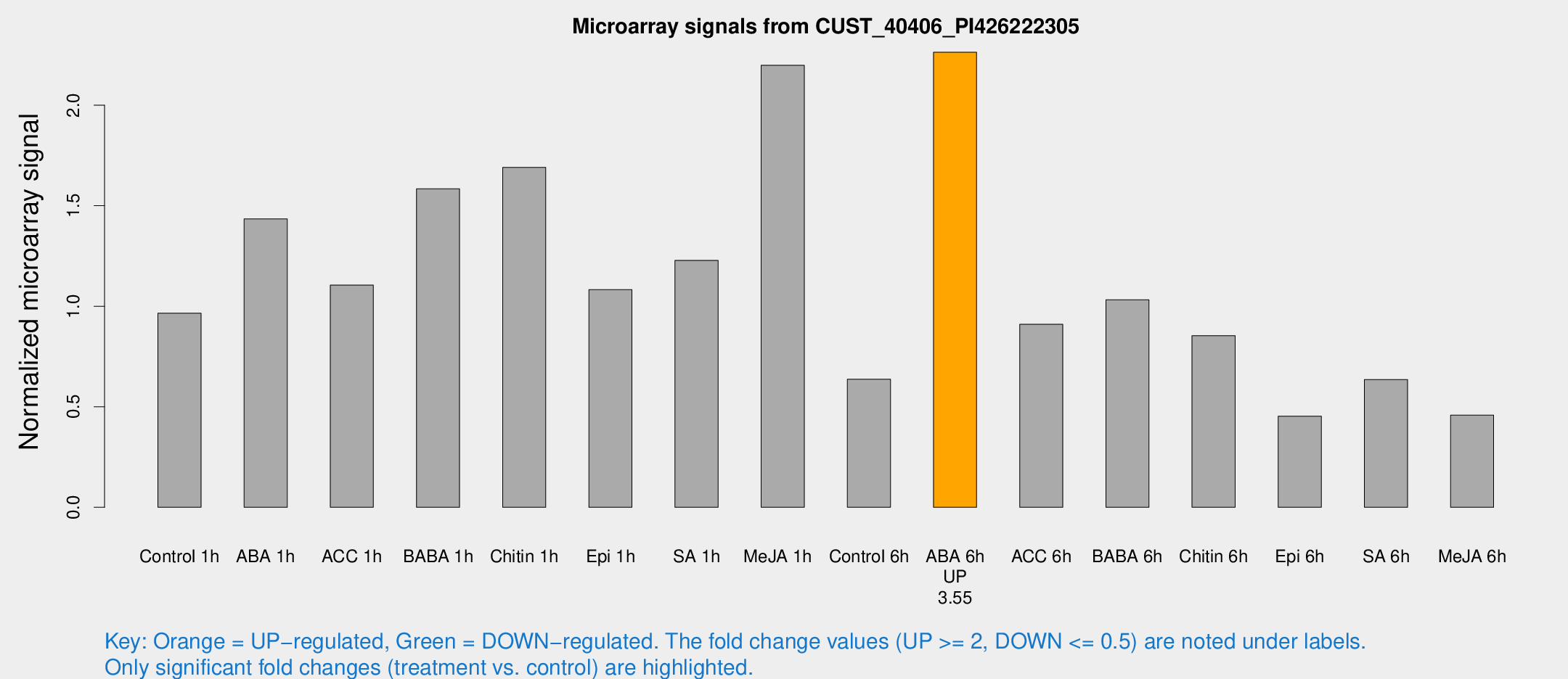
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 166.54 | 27.4062 | 0.965596 | 0.093806 |
| ABA 1h | 219.814 | 36.6669 | 1.43427 | 0.213324 |
| ACC 1h | 193.433 | 22.9721 | 1.1051 | 0.133518 |
| BABA 1h | 259.574 | 27.173 | 1.58416 | 0.110808 |
| Chitin 1h | 267.048 | 52.8936 | 1.69057 | 0.298385 |
| Epi 1h | 158.99 | 15.3085 | 1.08236 | 0.0748402 |
| SA 1h | 217.841 | 36.4323 | 1.22819 | 0.195838 |
| Me-JA 1h | 303.279 | 20.7901 | 2.19858 | 0.200747 |
| Control 6h | 123.905 | 39.1129 | 0.636972 | 0.219916 |
| ABA 6h | 415.881 | 67.5112 | 2.26352 | 0.455364 |
| ACC 6h | 177.39 | 17.7927 | 0.910195 | 0.0556599 |
| BABA 6h | 204.024 | 46.6702 | 1.03207 | 0.208518 |
| Chitin 6h | 153.48 | 9.4923 | 0.854021 | 0.0527243 |
| Epi 6h | 86.9248 | 8.76906 | 0.453441 | 0.031817 |
| SA 6h | 108.054 | 14.8953 | 0.635733 | 0.0419078 |
| Me-JA 6h | 79.5905 | 14.0702 | 0.458447 | 0.126414 |
Source Transcript PGSC0003DMT400061609 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
