Probe CUST_39791_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39791_PI426222305 | JHI_St_60k_v1 | DMT400062613 | GGTGTTTGGGAGTCTAAAAAATTGAGACATCTTTGTTACACAAAAGAATGTTACTGTGTC |
All Microarray Probes Designed to Gene DMG400024366
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39791_PI426222305 | JHI_St_60k_v1 | DMT400062613 | GGTGTTTGGGAGTCTAAAAAATTGAGACATCTTTGTTACACAAAAGAATGTTACTGTGTC |
Microarray Signals from CUST_39791_PI426222305
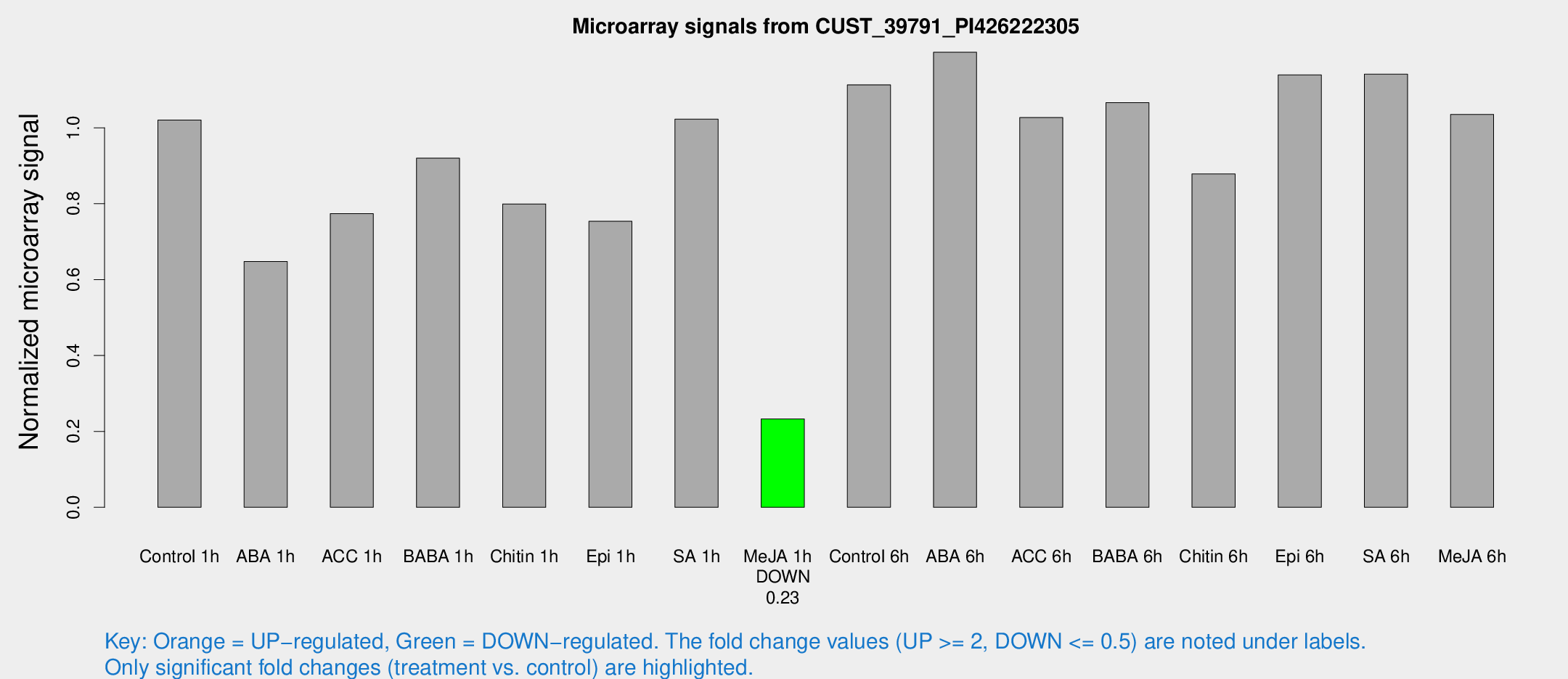
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 41.3979 | 4.01127 | 1.02044 | 0.0999944 |
| ABA 1h | 27.3606 | 9.48906 | 0.647659 | 0.280417 |
| ACC 1h | 40.6268 | 15.1584 | 0.7736 | 0.437282 |
| BABA 1h | 36.2321 | 4.27003 | 0.920016 | 0.10327 |
| Chitin 1h | 32.5478 | 10.4181 | 0.799333 | 0.290098 |
| Epi 1h | 27.0741 | 4.46269 | 0.75361 | 0.131551 |
| SA 1h | 43.9412 | 8.7715 | 1.02284 | 0.138456 |
| Me-JA 1h | 7.66273 | 3.32754 | 0.232648 | 0.101882 |
| Control 6h | 56.0762 | 21.0179 | 1.11314 | 0.559456 |
| ABA 6h | 52.7201 | 9.07446 | 1.19914 | 0.139652 |
| ACC 6h | 48.1441 | 6.11244 | 1.02711 | 0.106549 |
| BABA 6h | 50.5996 | 10.4533 | 1.06617 | 0.222537 |
| Chitin 6h | 40.0869 | 9.452 | 0.878319 | 0.251644 |
| Epi 6h | 51.9684 | 4.89157 | 1.13945 | 0.107596 |
| SA 6h | 47.2194 | 8.59312 | 1.14125 | 0.116788 |
| Me-JA 6h | 45.4067 | 11.9147 | 1.03522 | 0.258728 |
Source Transcript PGSC0003DMT400062613 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G58410.1 | +1 | 7e-27 | 111 | 82/286 (29%) | Disease resistance protein (CC-NBS-LRR class) family | chr1:21701286-21704255 REVERSE LENGTH=899 |
