Probe CUST_39561_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39561_PI426222305 | JHI_St_60k_v1 | DMT400048720 | GGGATACGATACTAAATTCATGGCAGCTGTTACTAAAATTCATGACATCGTTTAAGCTAA |
All Microarray Probes Designed to Gene DMG400018934
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39561_PI426222305 | JHI_St_60k_v1 | DMT400048720 | GGGATACGATACTAAATTCATGGCAGCTGTTACTAAAATTCATGACATCGTTTAAGCTAA |
| CUST_39563_PI426222305 | JHI_St_60k_v1 | DMT400048721 | GTTAATTTCTCCATAATTATTTATATCTATGGTCGTCAGTTTAGGGTTGTGGAACTGCCA |
Microarray Signals from CUST_39561_PI426222305
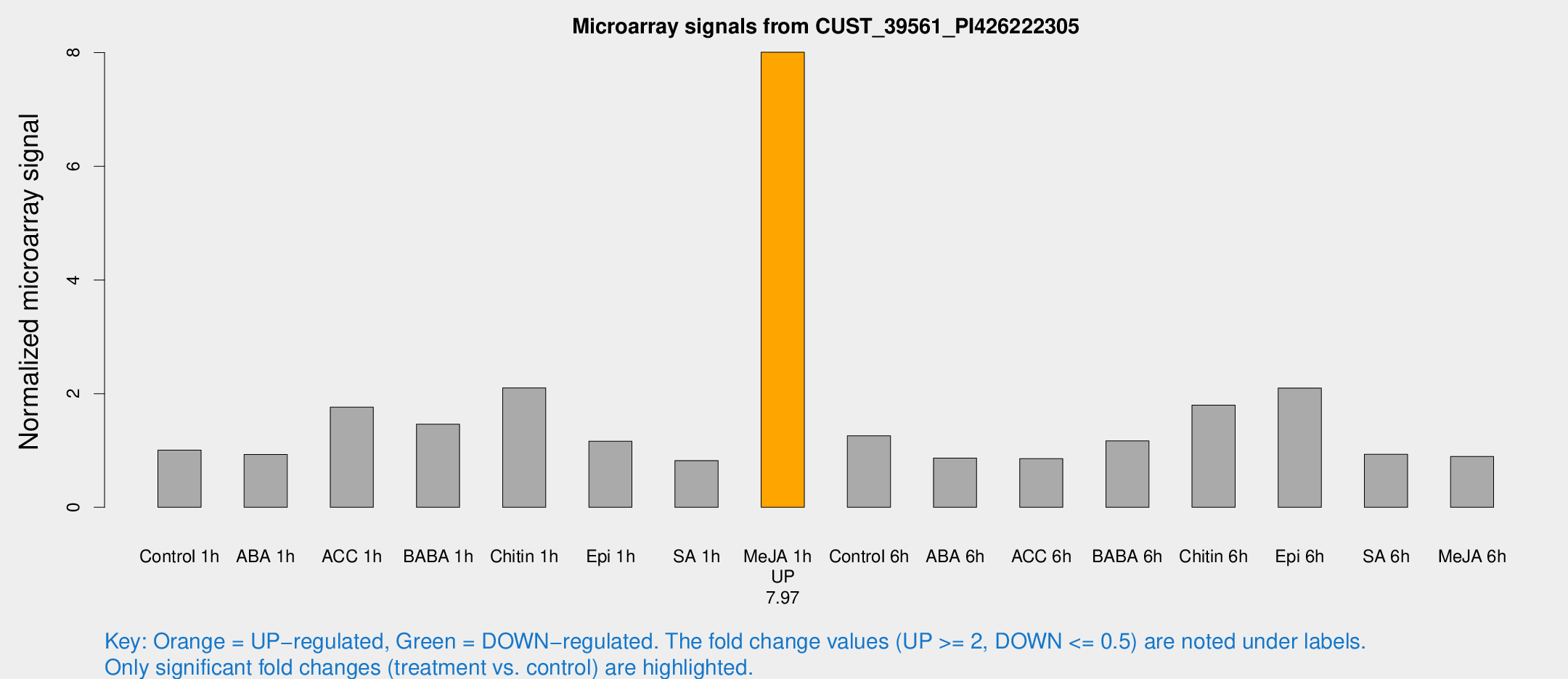
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.04226 | 2.84813 | 1.00481 | 0.471976 |
| ABA 1h | 4.73215 | 2.74168 | 0.931037 | 0.510286 |
| ACC 1h | 12.1718 | 3.32436 | 1.76224 | 0.756174 |
| BABA 1h | 9.46358 | 3.1605 | 1.46159 | 0.587578 |
| Chitin 1h | 21.5681 | 16.283 | 2.10145 | 2.75322 |
| Epi 1h | 6.19808 | 2.86076 | 1.1631 | 0.557558 |
| SA 1h | 5.10233 | 2.86511 | 0.821647 | 0.45964 |
| Me-JA 1h | 40.2532 | 4.58229 | 8.0067 | 1.67591 |
| Control 6h | 9.11459 | 4.0173 | 1.2573 | 0.580464 |
| ABA 6h | 5.61857 | 3.09129 | 0.867269 | 0.479076 |
| ACC 6h | 6.07668 | 3.59752 | 0.854901 | 0.495252 |
| BABA 6h | 8.39936 | 3.37647 | 1.16941 | 0.516004 |
| Chitin 6h | 12.6057 | 3.62404 | 1.79819 | 0.584482 |
| Epi 6h | 16.3934 | 5.28354 | 2.09819 | 1.06344 |
| SA 6h | 5.61509 | 3.14656 | 0.93206 | 0.52379 |
| Me-JA 6h | 5.37061 | 2.97966 | 0.895295 | 0.492692 |
Source Transcript PGSC0003DMT400048720 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G67150.1 | +2 | 3e-101 | 315 | 186/433 (43%) | HXXXD-type acyl-transferase family protein | chr5:26795880-26797226 REVERSE LENGTH=448 |
