Probe CUST_39517_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39517_PI426222305 | JHI_St_60k_v1 | DMT400019059 | AGTAATTTGATTTTTATGCAGCGCGAGACGTTGGATGAAGAATTGATTAGGATGAGAGAG |
All Microarray Probes Designed to Gene DMG400007387
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39514_PI426222305 | JHI_St_60k_v1 | DMT400019058 | AATAGTTTTTCCATTGACCTTAACACAAACCTCTCTTTGCACAACACCAATACAAGTCCG |
| CUST_39517_PI426222305 | JHI_St_60k_v1 | DMT400019059 | AGTAATTTGATTTTTATGCAGCGCGAGACGTTGGATGAAGAATTGATTAGGATGAGAGAG |
Microarray Signals from CUST_39517_PI426222305
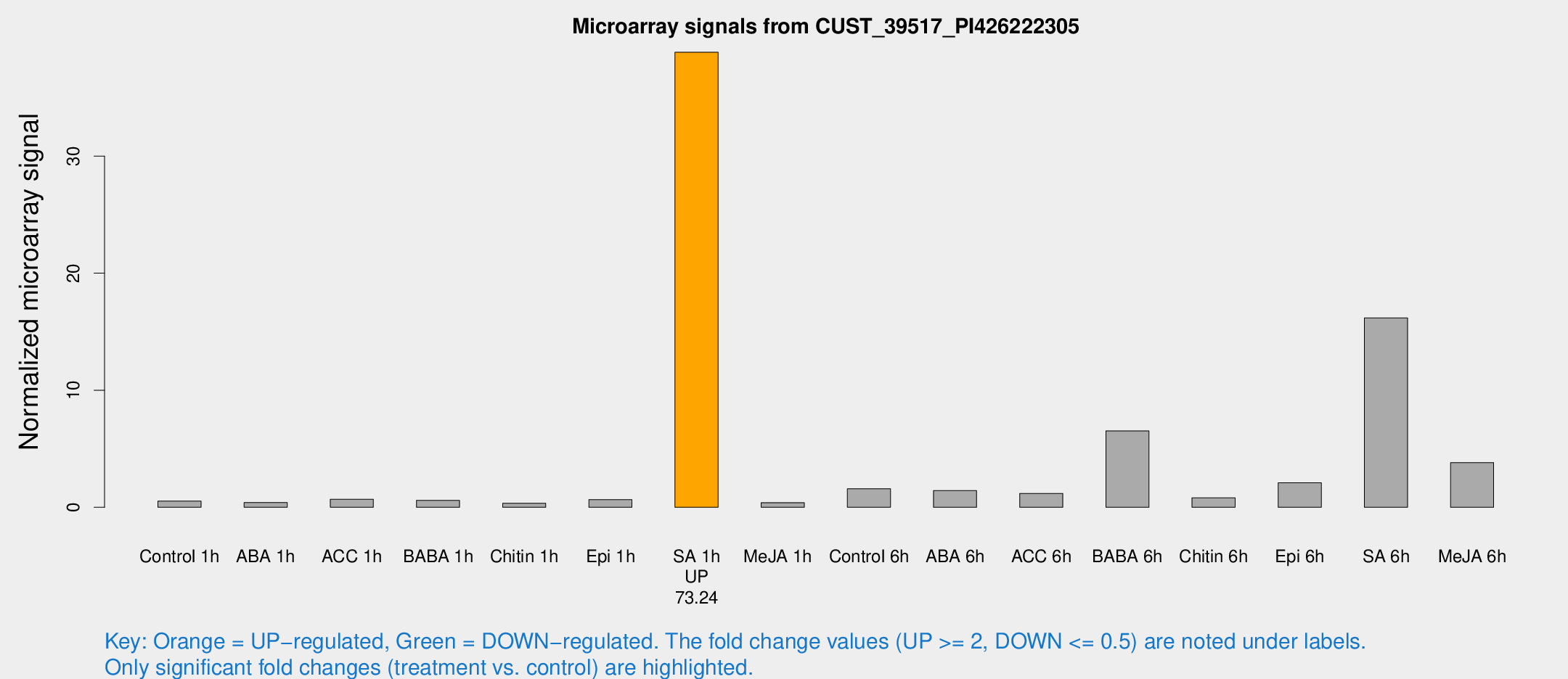
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 13.7148 | 4.73065 | 0.530862 | 0.213816 |
| ABA 1h | 9.07356 | 3.52913 | 0.41177 | 0.195845 |
| ACC 1h | 24.3697 | 15.943 | 0.680877 | 0.61752 |
| BABA 1h | 14.103 | 4.69451 | 0.592844 | 0.211576 |
| Chitin 1h | 7.00366 | 3.63174 | 0.347145 | 0.181418 |
| Epi 1h | 14.2294 | 4.24146 | 0.654917 | 0.247236 |
| SA 1h | 936.053 | 201.226 | 38.8784 | 6.30713 |
| Me-JA 1h | 7.27746 | 3.79954 | 0.39463 | 0.206189 |
| Control 6h | 44.1033 | 18.4333 | 1.5911 | 0.708458 |
| ABA 6h | 39.4877 | 12.6785 | 1.42645 | 0.618712 |
| ACC 6h | 31.5874 | 5.80437 | 1.18488 | 0.441416 |
| BABA 6h | 168.191 | 30.3735 | 6.52799 | 0.932419 |
| Chitin 6h | 23.2678 | 10.1417 | 0.81037 | 0.379271 |
| Epi 6h | 55.4562 | 11.2375 | 2.09507 | 0.346152 |
| SA 6h | 408.347 | 128.666 | 16.1846 | 9.38069 |
| Me-JA 6h | 126.795 | 75.7839 | 3.8166 | 3.15462 |
Source Transcript PGSC0003DMT400019059 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G80840.1 | +2 | 1e-60 | 197 | 125/271 (46%) | WRKY DNA-binding protein 40 | chr1:30383834-30385356 FORWARD LENGTH=302 |
